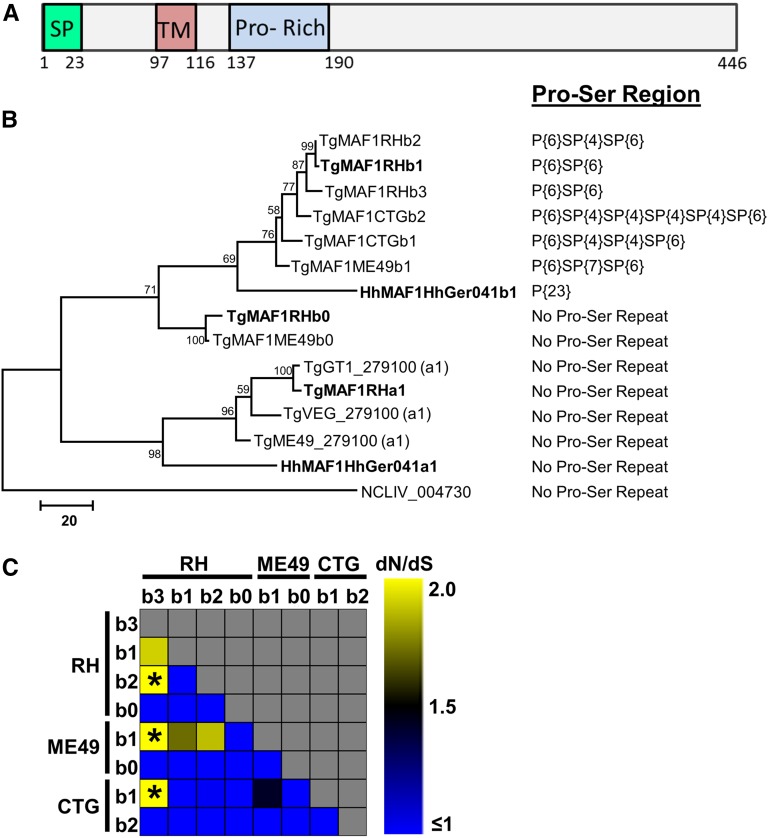
Figure 2.
The T. gondii and H. hammondi MAF1 loci harbor two distinct isoforms while only one isoform is present in N. caninum. (A) Schematic representation of the predicted MAF1 protein. The signal peptide (SP) was predicted using SignalP v4.0 and the putative transmembrane domain (TM) was predicted by TMHMM v2.0. The proline-rich region (Pro-Rich) stretches from AA152 to 164 of TgMAF1RHb1 and is not found within all MAF1 paralogs (e.g., TgMAF1RHa1, a2). (B) Phylogram of either cloned MAF1 amino acid sequences from T. gondii, H. hammondi, and N. caninum, or those downloaded directly from ToxoDB (with TG Gene nos.). Cloned sequences of all of the “b” paralogs from T. gondii did not match any predicted gene models in ToxoDB in terms of predicted coding region length and were left out of the analysis. Paralog family is indicated at the end of each name (e.g., a1, b1, b2, etc.) (C) dN/dS ratio calculations for all T. gondii “b” MAF1 paralogs, including b0. * indicates significant evidence for diversifying selection for that particular paralog comparison (P < 0.05).