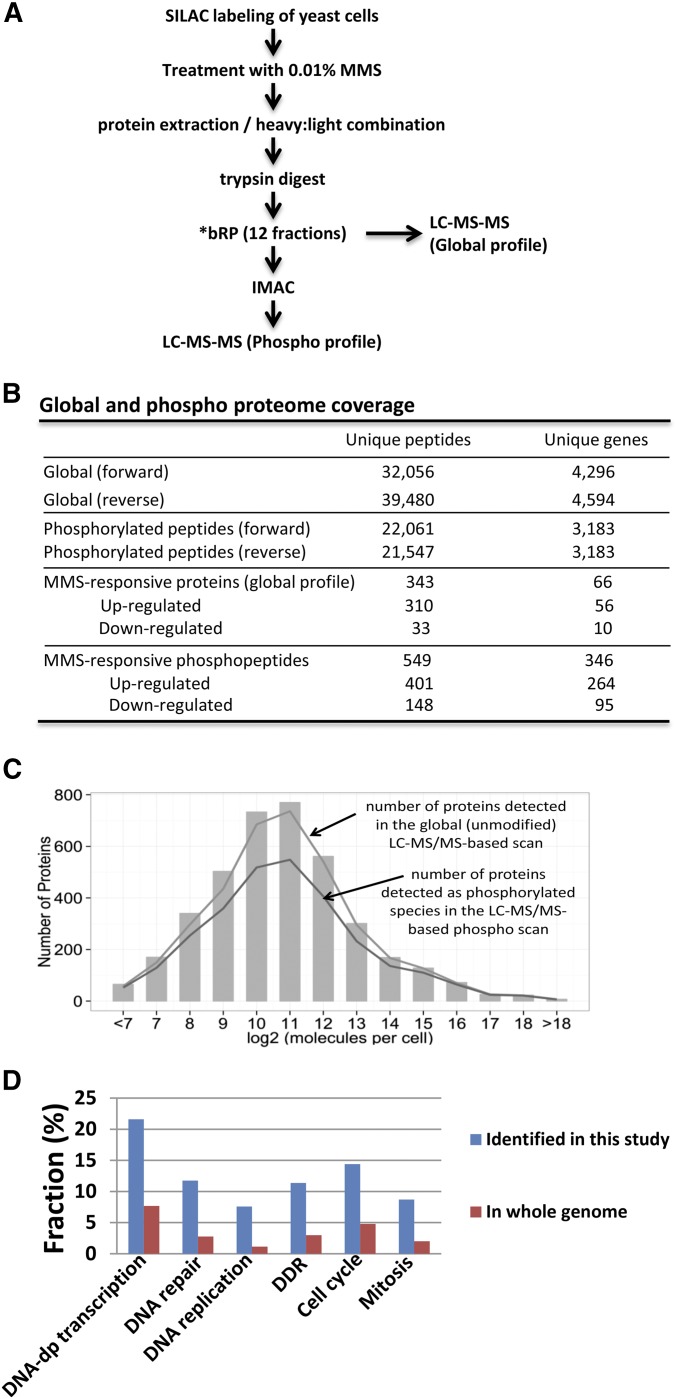
Figure 1.
Identification of MMS-responsive phospho-peptides by LC-MS/MS. (A) Strategy. We exposed heavy (H)-SILAC-labeled wild-type yeast cells to 0.01% MMS and mock-exposed light (L)-SILAC-labeled cells. After 3 hr, H- and L-labeled cells were mixed in equal amounts by protein mass. To ensure reproducibility, the entire experiment was repeated, and the labels were swapped such that the (L)-SILAC-labeled wild-type yeast cells were exposed to 0.01% MMS for 3 hr, and the (H)-SILAC-labeled wild-type yeast cells were mock-exposed. *bRP: basic reverse-phase (bRP) liquid chromatography (LC). (B) Global and phospho-proteome coverage. Note that “MMS-responsive” phospho-peptides include the peptides the phosphorylation of which increases (SILAC ratio ≥2) or decreases (SILAC ratio ≤0.5) after MMS treatment. (C) The high coverage of the yeast proteome achieved by LC-MS/MS is not biased. Bar graph is extracted from Ghaemmaghami et al. (2003) and represents the abundance distribution of 80% of the yeast proteome based on immuno-detection. Lines represent the abundance-based coverage achieved in our LC-MS/MS-based global and phospho scans. (D) The genes encoding MMS-inducible phosphoproteins are enriched in the functional categories of cell cycle regulation and DDR. Gene ontology enrichment analysis was conducted on the 264 MMS-induced phosphoproteins (≥2σ or twofold) using the Funspec software package (http://funspec.med.utoronto.ca/). After inputting the 264 gene names and setting the P-value cutoff as 10−6, the following processes were found to be enriched: DNA-dependent transcription (P = 1.1 × 10−13), DNA repair (P = 2.5 × 10−12), DNA replication (P = 4.1 × 10−12), DNA damage response (P = 9.2 × 10−11), cell cycle (P = 3.4 × 10−10), and mitosis (P = 4.1 × 10−9).