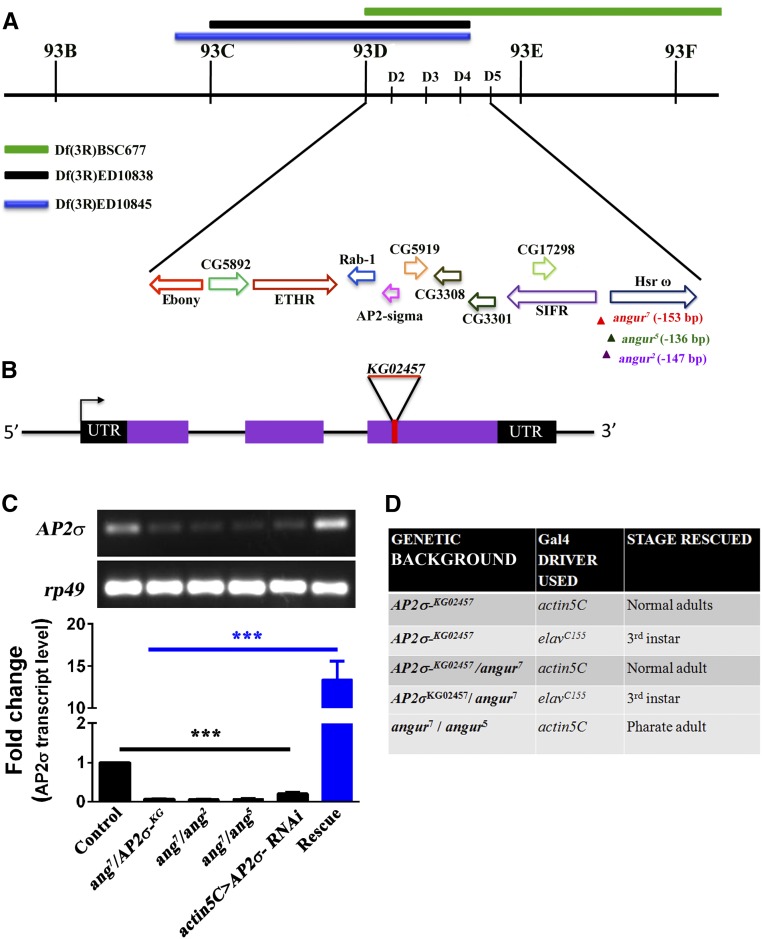
Figure 1.
angur is an allele of σ2-adaptin, the smallest subunit of adapter protein 2 (AP2) complex. (A) Deficiency mapping showed the angur locus to be located in the region of 93D1–93D4 on the third chromosome of Drosophila. The three deficiency lines that uncovered the angur locus—Df(3R)BSC677, Df(3R)ED10838, and Df(3R) ED10845—are labeled. Solid bars mark the deleted regions in these deficiency lines. The 93D1–93D4 region of the chromosome contains at least 10 identified genes. Different colored arrows represent the relative positions and orientations of the genes in the 93D1–93D4 region. While angur7 and angur2 were obtained from mobilization of EP0877, angur5 was obtained from mobilization of KG06118b. The P-element in angur alleles was located in the 5′ end of the hsrω gene but did not affect expression of the hsrω gene. (B) Genomic organization of the σ2-adaptin locus showing exons (represented by solid boxes) and introns (represented by thin lines). The transcription start site is represented by an arrow, and the untranslated regions are shown by black solid boxes. The insertion site of P-element KG02457 lies in the third exon of the gene. (C) Semiquantitative (top panel) and quantitative RT-PCR (bottom panel) depicting transcript levels of σ2-adaptin in controls, angur7/AP2σKG02457, angur7/angur2, angur7/angur5, actin5C-Gal4-driven σ2-adaptin RNAi, and rescued heteroallelic mutant (actin5C/+; angur7/UAS-AP2σ, AP2σKG02457), respectively. σ2-adaptin transcript level is dramatically reduced in the heteroallelic mutants or actin5C-Gal4-driven σ2-adaptin RNAi. rp49 transcript level was used as an internal concentration control for messenger RNA (mRNA). The error bars in the bottom panel represent SEM. Statistical analysis based on one-way ANOVA followed by post-hoc Tukey’s multiple comparison test. (D) Rescue of AP2σKG02457 mutants and heteroallelic combinations in homozygous or transheterozygous combinations using ubiquitous actin5C-Gal4 or the neuronal elavC155-Gal4 drivers.