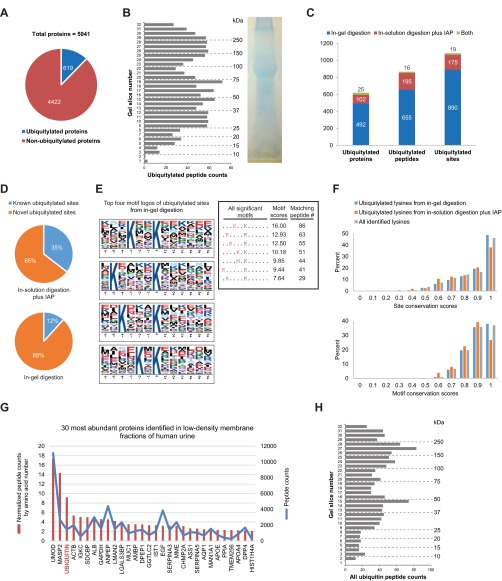
Fig. 3.
Large-scale identification of ubiquitylated proteins in urinary exosomes. (A) Total ubiquitylated and nonubiquitylated proteins identified in human urinary exosomes. (B) Quantity of ubiquitylated peptides from 1D SDS-PAGE gel. (C) Total ubiquitylated proteins, peptides, and sites identified from in-gel digestion, in-solution digestion with IAP, or both. (D) Novel versus known ubiquitylated sites identified from either in-solution digestion with IAP or in-gel digestion. (E) Top four motif logos of ubiquitylated sites identified from in-gel digestion display strong preferences for lysine at various positions N-terminal to the ubiquitylated lysine residue. Seven significant motifs with their scores and matching peptide numbers are shown in a table. The logos and scores of the overrepresented ubiquitylation motifs were generated by motif-x program. (F) Distributions of site and motif conservation scores of ubiquitylated lysines from in-gel digestion, in-solution digestion plus IAP, and all identified lysines. (G) Thirty most abundant proteins identified in low-density membrane fractions of human urine based on normalized and total peptide counts (see supplemental data 1 for the complete list). A gene symbol for each protein is displayed, except for ubiquitin (encoded by four different genes i.e. RPS27A, UBA52, UBB, and UBC). (H) Instances of ubiquitin peptide identification from 1D SDS-PAGE gel.