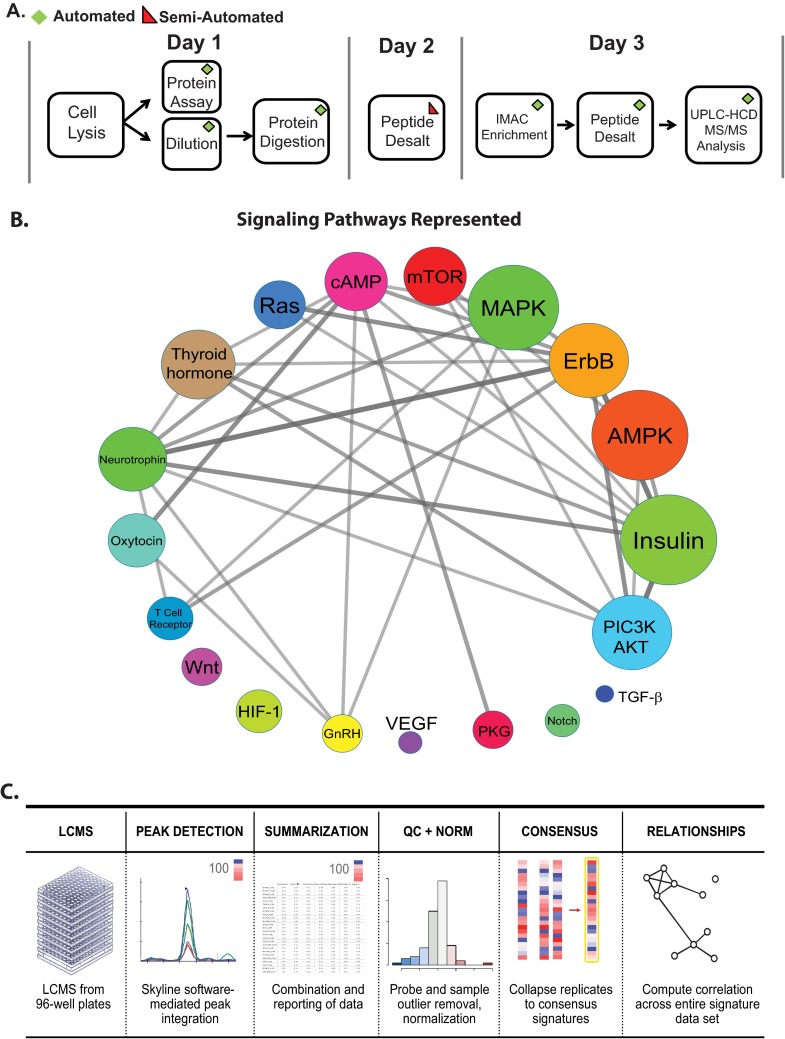
Fig. 3.
P100 automated sample processing, pathway coverage, and data analysis pipeline. A, A schematic of automated P100 sample processing. On the first day, cells are lysed, subjected to protein quantification and diluted to a uniform concentration. Proteins are then reduced, alkylated, and digested. On the second day, samples are desalted using a 96-well device and dried overnight. On the third day, phosphopeptide enrichment by IMAC and desalting occurs. Finally, samples are analyzed using high resolution UPLC-HCD MS/MS. B, The top signaling pathways represented by the larger set of phosphopeptides used to identify the reduced-representation phosphopeptide probe set are shown. Each signaling pathway is depicted as a circle that is sized to indicate the number of source proteins involved in a specific pathway. A larger size indicates that a signaling pathway is represented by a larger number of phosphopeptides. The color of each pathway is only meant to show the diversity of the signaling pathways represented. C, The data analysis pipeline for the P100 assay is shown. Data are collected in a 96-well plate format, analyzed within Skyline, and exported for summarization. Phosphopeptide probes and sample outliers are removed and the light/heavy peptide ratios are normalized. Quality controlled data are hierarchically clustered and molecular signatures for different perturbations are revealed.