FIGURE 4.
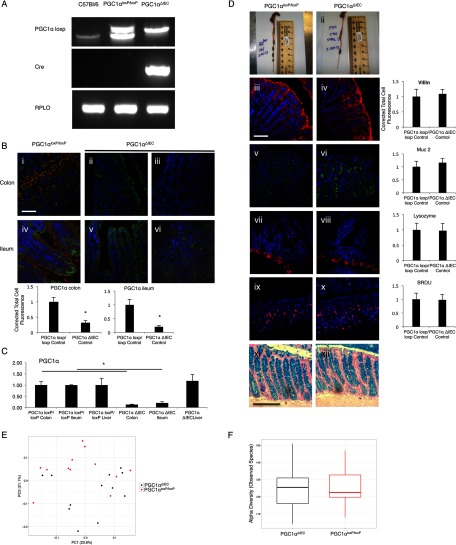
Intestinal epithelium-specific PGC1α knock-out mouse (PGC1αΔIEC) was created by mating a PGC1αloxP/loxP mouse with a villin-cre mouse. Mice were genotyped using RT-PCR (A). Immunofluorescence for PGC1α (Cy3, red), E-cad (FITC, green), and nuclear stain (DAPI, blue) demonstrates a significant decrease of PGC1α protein within the intestinal epithelium of PGC1αΔIEC mice (B, n = 8/group, scale bars, 100 μm). Staining in both the colon and ileum was quantified. Representative images are provided from the colon of PGC1αloxP/loxP mice (B, panel i) and PGC1αΔIEC mice (B, panels ii and iii) as well as the ileum of PGC1αloxP/loxP mice (B, panel iv) and PGC1αΔIEC mice (B, panels v and vi). qRT-PCR of intestinal tissue demonstrates PGC1α expression in the colon, ileum, and liver of PGC1αΔIEC and PGC1αloxP/loxP mice. Expression is decreased in the intestines of PGC1αΔIEC mice as compared with PGC1αloxP/loxP mice but not in the liver (C). No differences in intestinal morphology were noted between strains (D, panels i and ii). Immunofluorescence was performed for colonic villin (D, panels iii and iv, scale bars, 100 μm), colonic Muc2 (D, panels v and vi), ileal lysozyme (D, panels vii and viii), and colonic BrdU (D, panels ix and x). When quantification was performed, no differences were noted between strains. Alcian blue staining for goblet cells was similar between strains (D, xi and xii). β diversity comparisons of microbial communities within fecal samples from PGC1αΔIEC mice and their PGC1αloxP/loxP littermates are not significant. Shown are the observed species indices for each sample group (E). Displayed are principal coordinate analyses of unweighted Unifrac distances between all samples analyzed. Axis labels indicate the proportion of variance explained by each principal coordinate axis. Fecal samples of the two strains do not cluster separately within the principal coordinate space. Diversity within each group is as great as the diversity between sample types. α diversity comparisons of microbial communities within fecal samples from PGC1αΔIEC mice and their PGC1αloxP/loxP littermates are not significant. Shown are the observed species indices for each sample group (F). *, p ≤ 0.05.