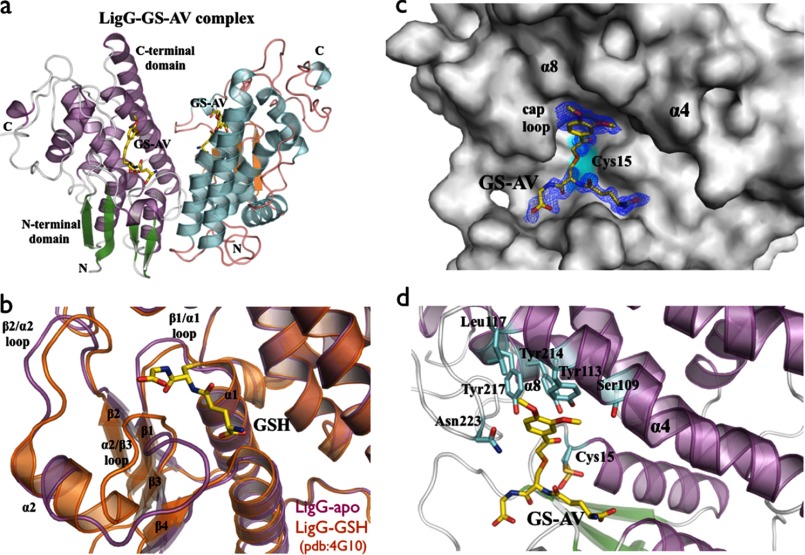
FIGURE 4.
a, overall schematic representation of the LigG·GS-AV complex dimer. b, superposition of the GSH binding site of apo-LigG (magenta) and LigG-GSH (Protein Data Bank entry 4G10) (orange) structures (30). Significant conformational changes of the GSH binding site were observed on the loop regions at the N-terminal domain connecting the β1/α1, β2/α2, and α2/β3 structural elements. c, molecular surface representation of the LigG monomer in complex with the GS-AV substrate analog. A feature-enhanced map (35) contoured at 1.0 σ is shown in blue around the GS-AV substrate analog (this molecule was omitted from the model to reduce bias). The position of the catalytic Cys15 residue is highlighted in cyan. d, active site of the LigG·GS-AV complex. The glutathionyl moiety of the GS-AV substrate sits on the top of the four β-strands of the N-terminal thioredoxin domain. In addition, the AV moiety of the GS-AV molecule contacts the C-terminal α-helical domain of LigG via residues Ser109, Tyr113, and Leu117 on α4, residues Tyr214 and Tyr217 on α8, and Asn223 on the cap loop region composed of the residues 220GGGNG224.