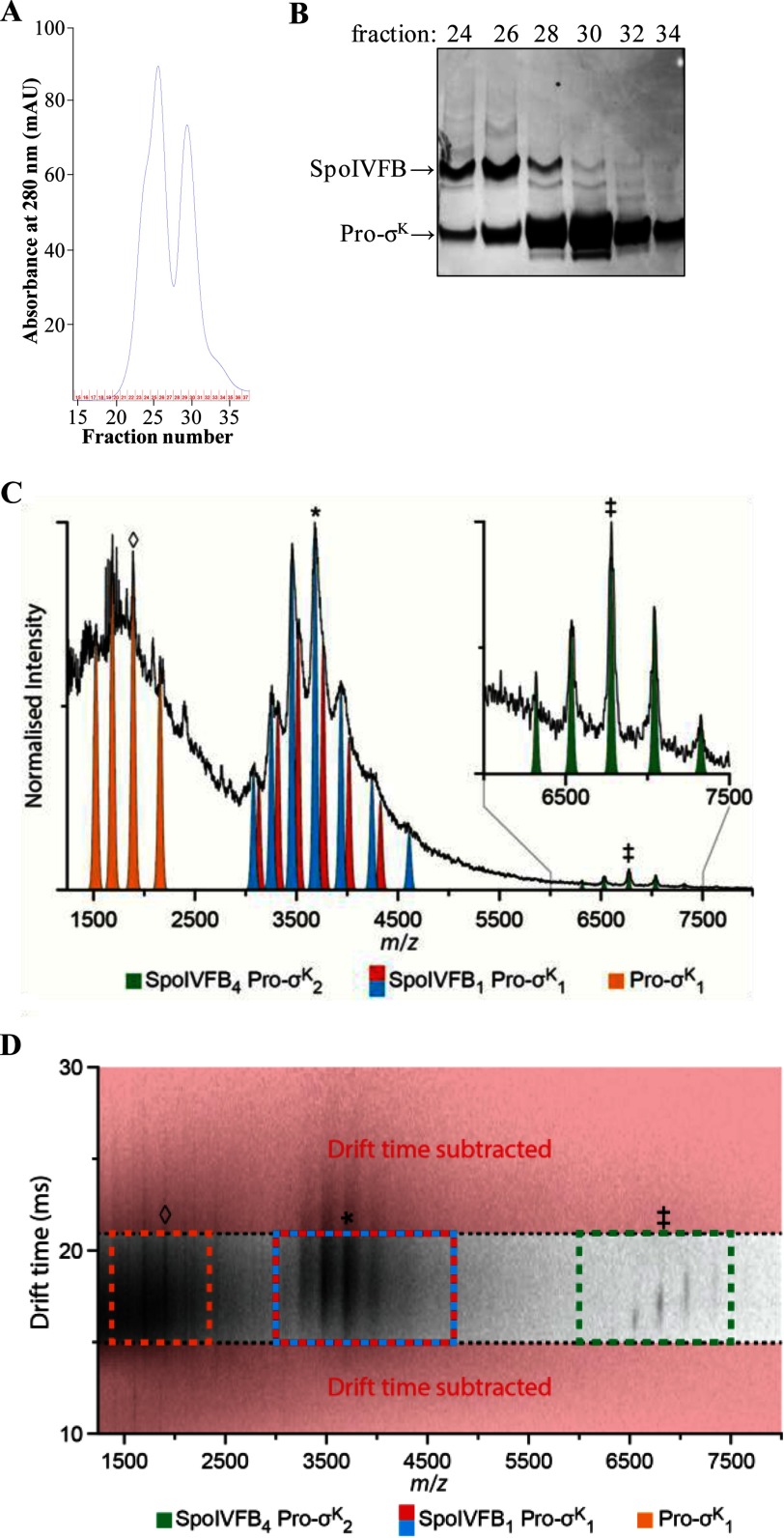
FIGURE 6.
Purification of the SpoIVFB-TEV-FLAG2 E44Q·Pro-σK(1–126)-His6 complex and native IM-MS analysis. A, gel filtration chromatography. The bound fraction from the cobalt affinity purification was concentrated and applied to a size exclusion column. The absorbance at 280 nm was monitored as fractions were collected, generating the profile shown. B, samples of fractions from the size exclusion column were subjected to SDS-PAGE as described for immunoblot analysis, but in this case gel electrophoresis was followed by Coomassie Blue staining of proteins. C, analysis of the SpoIVFB-TEV-FLAG2 E44Q·Pro-σK(1–126)-His6 complex prepared in a buffer containing 0.2% DDM revealed a mass of 183.2 kDa (green; ‡ = +27), agreeing well with a stoichiometry of 4:2 (SpoIVFB4 Pro-σK2). Applying post-IM CID methods (drift time axis is presented in D), three mobility time-aligned dissociation products were observed: monomeric Pro-σK(1–126)-His6 (Pro-σK1, orange; ♢ = +8) and two masses closely agreeing with a 1:1 complex (SpoIVFB1 Pro-σK1, blue and red; * = +15). The mass difference between these two products (blue, 55.3 kDa; red, 56.6 kDa) suggests that some lipid or detergent molecules remain bound in the complex. D, post-IM CID analysis. To enable the stoichiometry assignment of the complex, drift time subtraction was used to remove those ions not aligning with the antecedent precursor ions (red). Drift time subtraction was performed post-process using software packages outlined under “Experimental Procedures.” SpoIVFB4 Pro-σK2 (green), ‡ = +27; SpoIVFB1 Pro-σK1 (blue and red), * = +15; Pro-σK1 (orange), ♢ = +8. In A–D, representative results from two technical replicates of the purification and IM-MS analysis are shown.