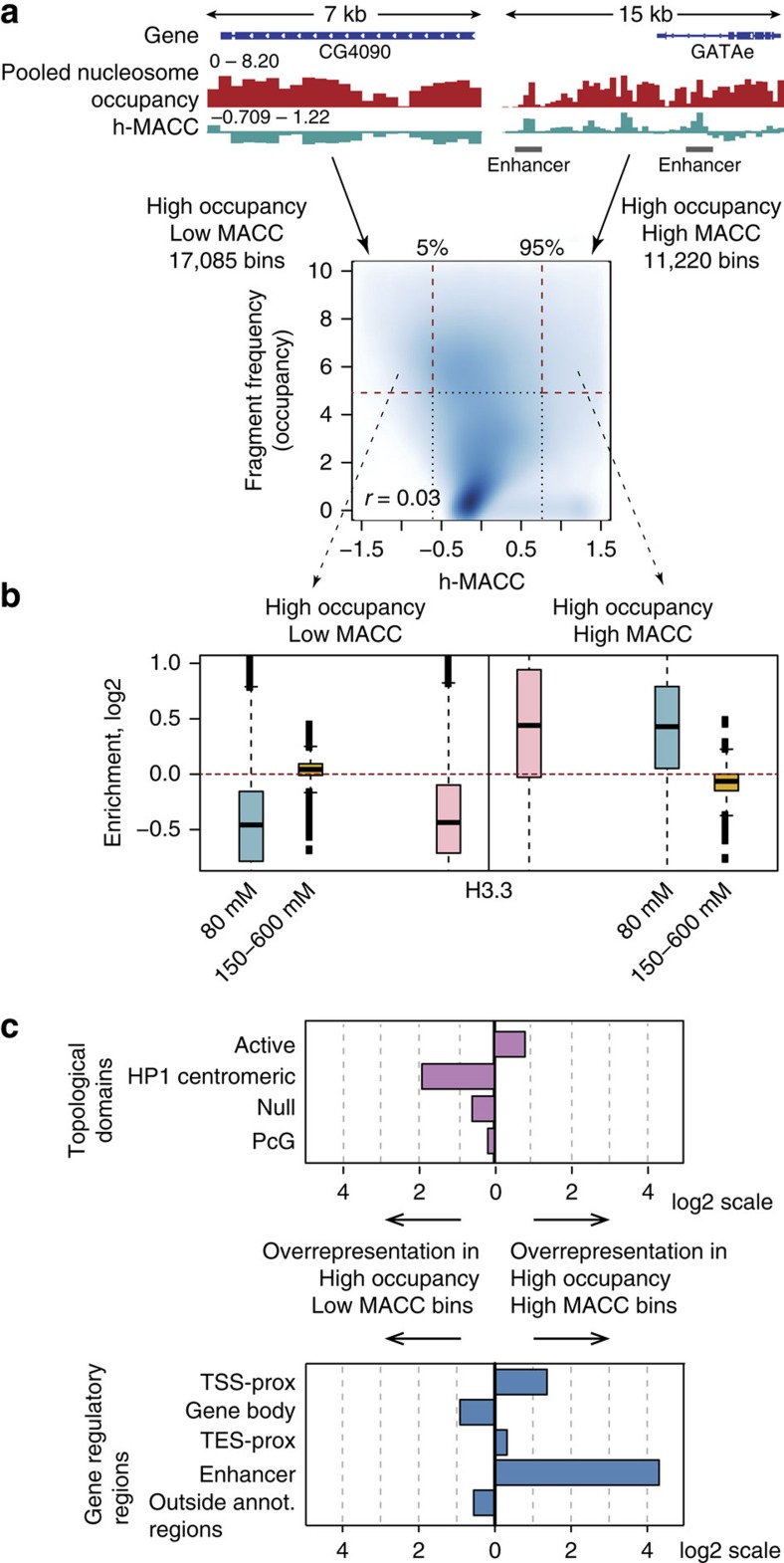
Figure 4. MACC is a better predictor of DNA accessibility than nucleosome occupancy.
(a) Examples (upper panels) and scatterplot (lower panel) showing association between MACC and ‘pooled' nucleosome occupancy. A single point on the scatterplot corresponds to one 300-bp bin (all bins for which MACC was estimated were included in this analysis). The Pearson's correlation coefficient between MACC and nucleosome occupancy is shown at the bottom. The dashed horizontal line indicates the high nucleosome occupancy cutoff (top 20% of bins), and the vertical dashed lines indicate MACC value thresholds for top and bottom 5% of bins. The numbers of genomic bins in each region are indicated on the plot. (b) Enrichment of salt-extracted chromatin fractions and H3.3 histone variant in the high occupancy bins shown in a. The 80 mM ‘active nucleosome' fraction is blue, the 150–600 mM ‘stable' or ‘repressed' fraction is orange, and the H3.3 enrichment is pink. The boxplots on the left and right sides of the figure (separated by the vertical black line) correspond to the bins with low and high MACC values respectively. Enrichment is plotted on a log2 scale and red horizontal line is placed at 0. (c) Comparison of the enrichment of two selected sets of bins in topological domains (upper panel, purple) and annotated genomic regions (lower panel, blue): TSS proximal (±1 kb), gene body, TES-proximal (±1 kb), and enhancers. For each bin set we calculated the fraction of bins overlapping a given region and the ratio of these fractions is shown on the plot.