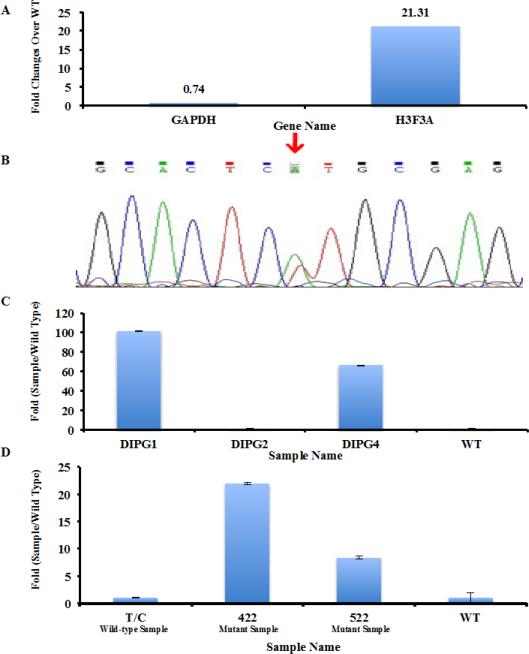
Fig. 5.
Detecting the H3F3A K27M mutation in DIPG samples with unknown H3F3A status. (a) Detection of H3F3A K27M mutation in a newly diagnosed DIPG case using qPCR. Amplification of a tumor sample with wild type H3F3A was used as control. The difference in amplification of house-keeping gene GAPDH and H3F3A between the wild type and test sample were shown. 1ng of cDNA library was used. (b) Sanger Sequencing of H3F3A cDNA isolated from the DIPG cancer was used to validate the presence of H3F3A K27M mutation. (c) Analysis of H3F3A K27M mutation in three additional DIPG samples. The mutation status of each sample was kept unknown to RZ. The PCR conditions established in Supplemental Fig. 2 and 10 ng of cDNA libraries were used. (d) Detecting the H3F3A K27M mutation in paraffin-embedded samples. 10 ng of cDNA libraries from three different paraffin samples were analyzed by real-time PCR.