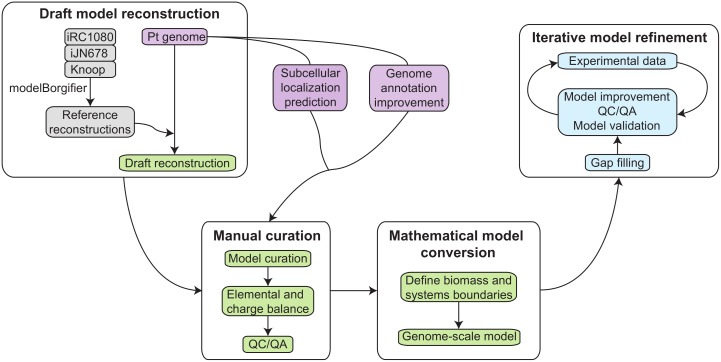
Fig 1. Metabolic network reconstruction workflow.
In step one we obtained a draft reconstruction based on P. tricornutum’s genome annotation and reference reconstructions. This draft reconstruction was manually curated using several resources such as an improved genome annotation, subcellular localization predictions and external databases. All reactions were elementally and charge balanced, QC/QA was performed and a biomass objective function was defined before transforming the reconstruction into a computational model. In an iterative process, the in silico predictions are compared with experimental observations to validate and improve the metabolic model.