FIG 1.
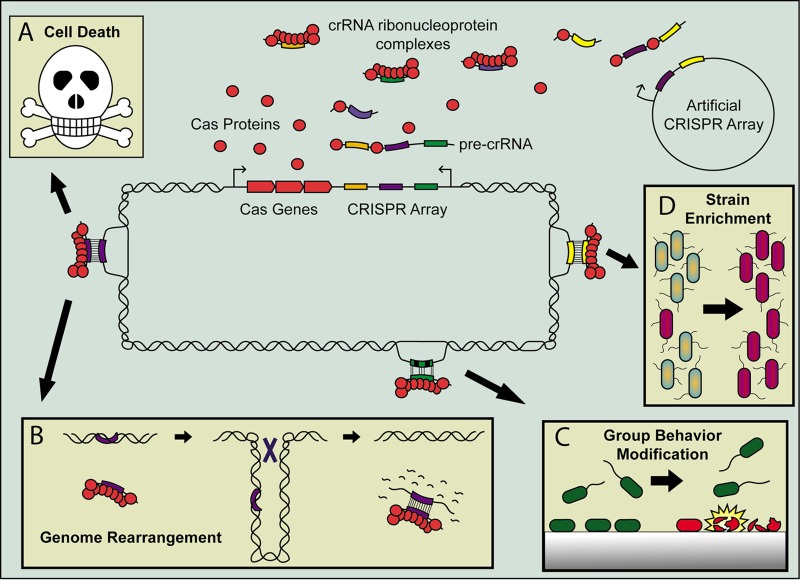
Consequences of CRISPR self-targeting in the type I system. The self-targeting crRNA can be transcribed from either the native CRISPR array or an artificial array on a plasmid, and the crRNA associates with the Cas proteins expressed from the native Cas-encoding operon to form the self-targeting crRNA ribonucleoprotein complex. These self-targeting spacers include both 100% complementary spacers (purple and yellow), as well as partially matching spacers (green and black). The most likely outcome of a 100% complementary match is cell death (A) due to the nucleolytic activity of the CRISPR-Cas system; however, this results in strong selection for mutants that remove the targeted region, for example, by genome rearrangement, such as excision of pathogenicity island or curing of a prophage (B). A partially matching self-targeting spacer can modulate group behavior, such as biofilm formation, by triggering viable planktonic cells (green cells) to die (red cells) upon surface binding (C). Bacterial populations transformed with a plasmid containing an artificial self-targeting spacer can enrich for bacterial strains that do not contain the target on their genome (purple cells, D).