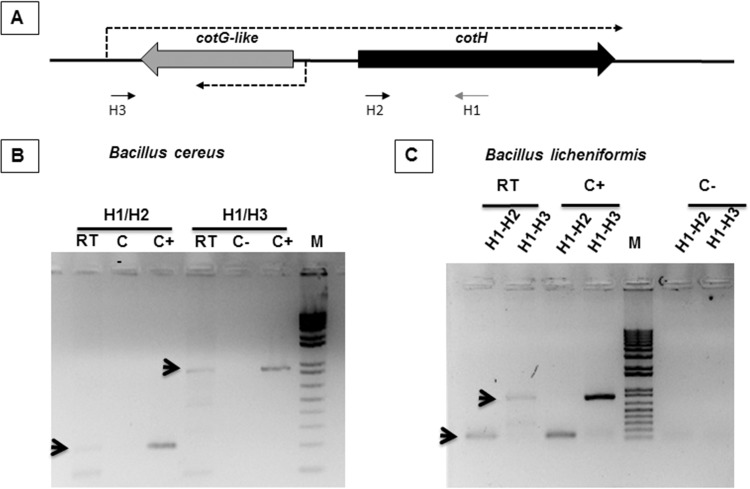
FIG 3.
cotH transcription in B. cereus and B. licheniformis. (A) Schematic representation of the cotH-cotG locus. Arrows indicate the positions of the synthetic oligonucleotides used for RT-PCR. Dashed arrows indicate the direction of transcription. (B and C) Reverse transcription reactions were performed by using total RNA from sporulating cells of B. cereus (B) or B. licheniformis (C) as a template and were primed with oligonucleotide H1. Amplification reactions were performed by using cDNA as the template and oligonucleotide pair H1/H2 or H1/H3, as indicated. Negative controls (C−) and positive controls (C+) were RNA samples treated and not treated with DNase, respectively. Arrows indicate the amplification products of the expected size, and M indicates the molecular weight marker.