FIG 1.
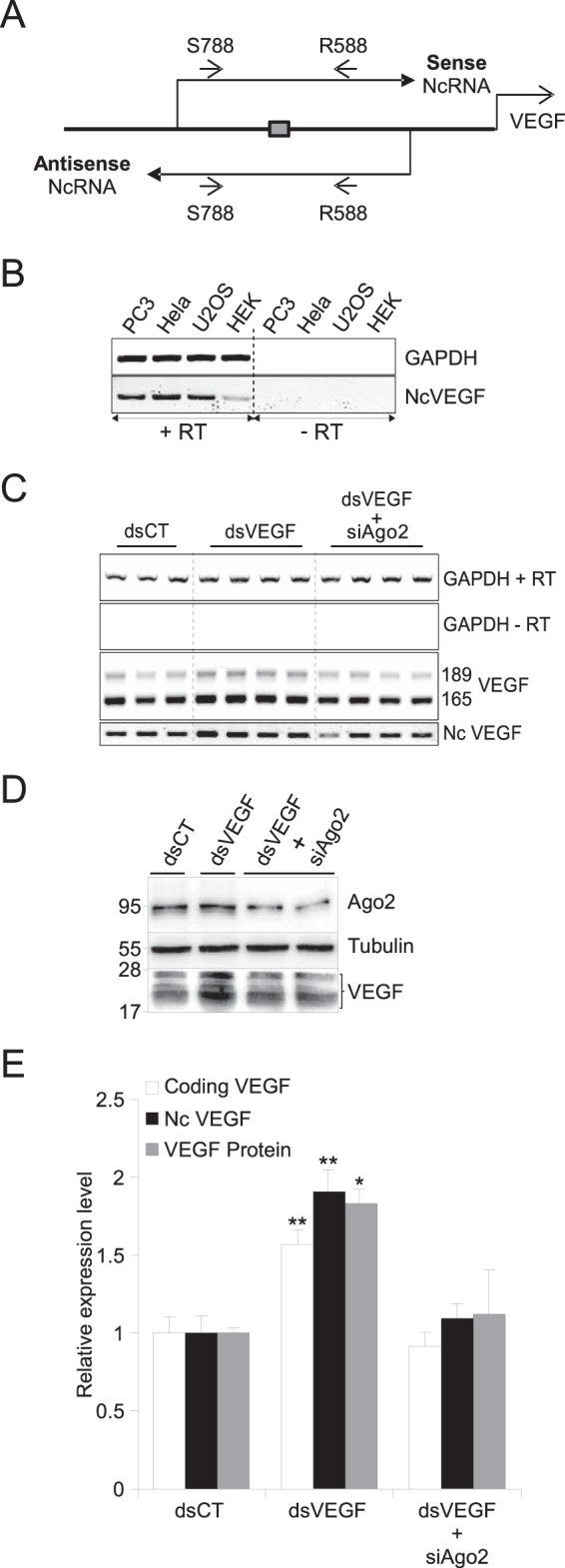
dsVEGF706 induces both VEGF and NcVEGF expression in an Ago2-dependent manner. (A) Schema of the strategy to identify sense and antisense NcVEGF RNA covering the dsVEGF706 target region (box). Small arrows represent sense qPCR-VEGF-S788 (S788) and antisense qPCR-VEGF-R588 (R588) primers listed in Table 2. (B) RNAs from PC-3, HeLa, U2OS, and HEK-293 cells were used in standard RT-PCR experiments with (+RT) or without (−RT) reverse transcriptase, followed by a PCR with the S788 and R588 primers. GAPDH amplification was used as a positive control. (C) Representative RT-PCR with RNA from PC-3 cells transfected with either control dsRNA (dsCT) or dsVEGF706 (dsVEGF) for 72 h without or with a set of siRNA targeting Ago2 (siAgo2). Transfections were processed in triplicate and quadruplicate. Both VEGF and NcVEGF expression levels were assessed by standard RT-PCR with (+RT) or without (−RT) reverse transcriptase. GAPDH served as a loading control. (D) Representative Western blot of PC-3 transfection experiments as described in panel C with anti-Ago2 and anti-VEGF antibodies. Tubulin was used as a loading control. (E) Histogram representing both qPCR and Western blot analyses of the transfection experiments described for panels C and D, respectively. The qPCR VEGF primers were designed to amplify only one fragment from either VEGF189 and VEGF165 cDNA and are not the same as the primers used in panel C (Table 2). 36B4 and GAPDH were used for normalization in qPCR experiments. Values are means ± the SD of three independent experiments, with each sample run in triplicate or quadruplicate. *, P ≤ 0.05; **, P ≤ 0.005.
