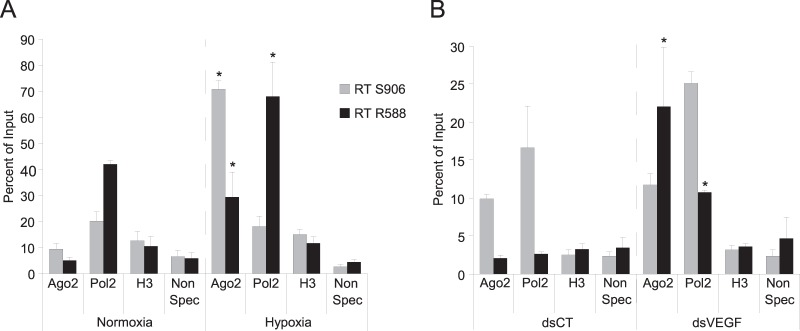
FIG 3.
Ago2 is recruited on the NcVEGF RNA in both hypoxic and dsVEGF706 transfected cells. PC-3 cells exposed either to normoxia/hypoxia (A) or dsCT/dsVEGF706 transfections (B) were used for RIP experiments. (A) RIP assays were performed on PC3 cells exposed to either hypoxia or normoxia using antibodies to Argonaute 2 (Ago2), RNA polymerase 2 (Pol2), acetyl-histone H3 (H3), or rabbit nonspecific serum (Non Spec) to pulldown-associated RNAs from PC3 cells in hypoxia or normoxia. The precipitated RNAs, as well as undiluted RNA input (10% RNA isolated from the supernatant of rabbit nonspecific serum IP), were used in gene-specific RT with either qPCR-VEGF-S906 or qPCR-VEGF-R588 primers, followed by a qPCR with specific primers surrounding the dsVEGF706 target region (S788-R706 [Table 2]). The results are reported as a histogram depicting the percent relative abundance of either the antisense (RT S906) or the sense (RT R588) NcVEGF compared to the total RNA input. (B) RIP assays were performed with the same antibodies, methods, and primers as in panel A, with pulldown-associated RNAs in PC3 cells transfected using either dsCT or dsVEGF706. The results are reported as a histogram depicting the percent relative abundance of either the antisense (RT S906) or sense (RT R588) NcVEGF compared to the total RNA input. Values are means ± the SD of three independent experiments with each sample run in triplicate. *, P ≤ 0.05.