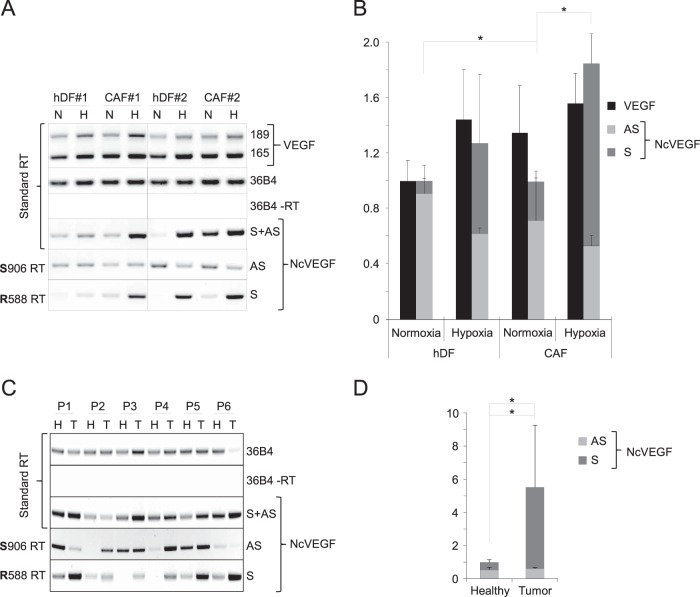
FIG 7.
Sense NcVEGF expression is increased in carcinoma-associated fibroblasts and in tumors. (A) Representative standard and gene-specific (R588 or S906) RT-PCR using RNAs from human primary dermal fibroblasts, hDF 1 and hDF 2, and carcinoma-associated fibroblasts, CAF 1 and CAF 2, exposed for 24 h to either normoxia (N) or hypoxia (H) as described in Materials and Methods. (B) cDNAs from these hDF and CAF fibroblasts were used in qPCR. The results are presented as a histogram depicting VEGF and the relative abundances of both antisense (S906) and sense (R588) NcVEGF in normoxia or hypoxia. The results are reported as the means ± the SD, with each sample run in triplicate. 36B4 was used to normalize qPCR experiments. *, P ≤ 0.05. (C) Representative standard and gene-specific (R588 or S906) RT-PCR using RNAs of either healthy (H) or tumor (T) lung tissues from six patients (P1 to P6 [Table 4]). The respective expressions of the sense (R588), antisense (S906), or both (standard RT) NcVEGF transcripts were revealed with NcVEGF primers (S788; R588 or S788; R706 [data not shown]). Standard RT-PCR with (+RT) or without (−RT) reverse transcriptase for 36B4 and GAPDH (data not shown) amplification were used as positive and negative controls. Experiments were processed in duplicate. (D) cDNAs from these six patients (P1 to P6) were used in qPCR. The results are presented as a histogram depicting the relative abundance of antisense (S906) and sense (R588) NcVEGF in each healthy sample and in corresponding tumor lung tissues. The results are reported as means ± the standard errors of the mean, with each sample run in triplicate. 36B4 and GAPDH were used for normalization in qPCR experiments. *, P ≤ 0.05.