FIG 3.
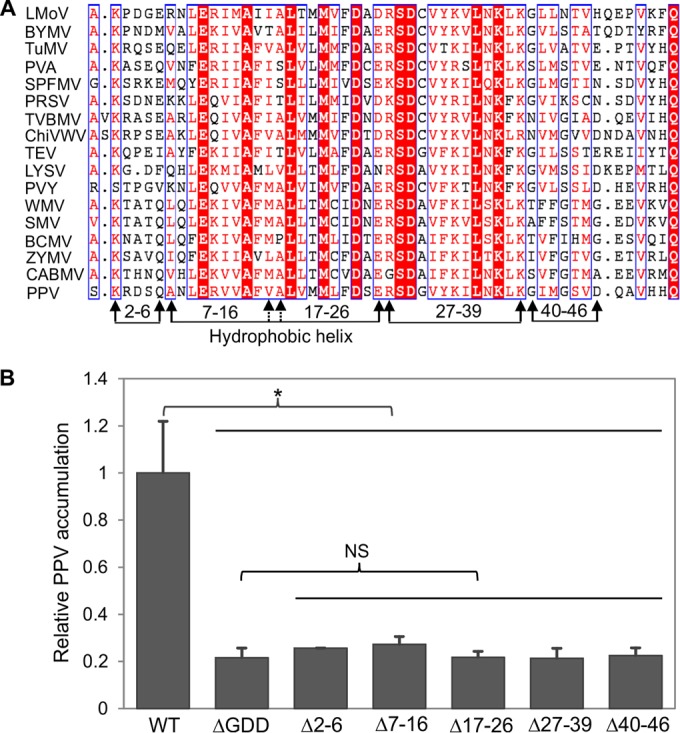
Evaluation of different motifs/regions of PPV 6K1 contributing to viral viability. (A) A multiple-sequence alignment of the 6K1 protein sequences of 17 potyviruses. Except for the 6K1 sequence of the PPV VPH isolate obtained from this study, the sequences of the other isolates were retrieved from the NCBI GenBank database. The sequences of the following viruses (GenBank accession numbers) are shown: Lily mottle virus (LMoV; AB570195), Bean yellow mosaic virus (BYMV; JX173278), Turnip mosaic virus (TuMV; AF169561), Potato virus A (PVA; AJ131402), Sweet potato feathery mottle virus (SPFMV; AB465608), Papaya ringspot virus (PRSV; X97251), Tobacco vein banding mosaic virus (TVBMV; EU734432), Chilli veinal mottle virus (ChiVWV; AJ972878), Tobacco etch virus (TEV; M11458), Leek yellow stripe virus (LYSV; AB194622), Potato virus Y (PVY; AJ439544), Watermelon mosaic virus (WMV; EU660581), Soybean mosaic virus (SMV; AF241739), Bean common mosaic virus (BCMV; AY863025), Zucchini mosaic virus (ZYMV; AF014811), and Cowpea aphid-borne mosaic virus (CABMV; HQ880242. The hydrophobic helix domains of potyviral 6K1 proteins are shown, and the amino acids are numbered according to the PPV 6K1 sequence. The different motifs/regions subjected to deletion analyses are indicated by arrows. Identical residues are shown as white letters in red background, whereas conserved substitutions are colored in red in blue boxes. (B) Detection of viral RNA accumulation in N. benthamiana plants agroinfiltrated with PPV mutants, the WT, and the nonreplicating ΔGDD mutant controls using qRT-PCR. Cells from each agrobacterial culture (OD600, 0.1) harboring the corresponding plasmid were infiltrated into nine uniform N. benthamiana plants at the 6- to 7-leaf stage. Agroinfiltrated leaves were collected and pooled at 54 hpi for RNA purification. Real-time qPCR was performed to quantify viral genome accumulation by analysis of the CP RNA level using the mCherry transcript level as an internal control. Error bars denote standard errors from three biological replicates. Statistically significant differences, determined by an unpaired two-tailed Student's t test, are indicated by brackets. NS, no significant differences; *, P < 0.05.
