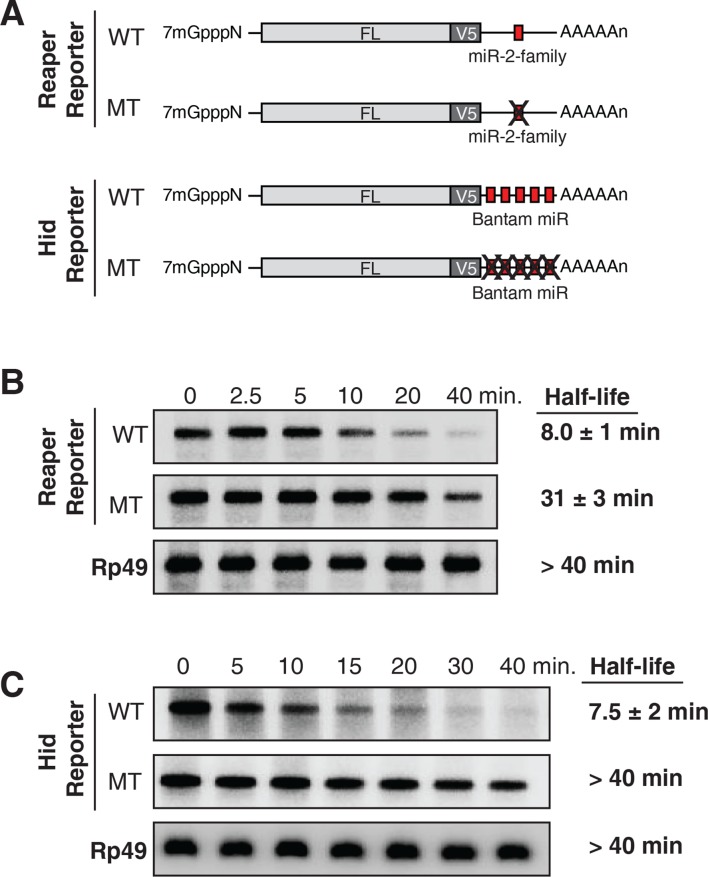
Figure 1. Destabilization of reaper and hid reporter mRNAs by endogenous miRNAs.
(A) Schematic of reporter mRNAs analyzed; WT denotes mRNAs with wild type MREs and MT denotes mRNAs where the MREs were inactivated by mutation. FL is the firefly luciferase coding region and V5 is the V5 epitope tag. (B) Stability of reaper reporter mRNAs containing wild type (WT) or mutant (MT) MREs. Cells were treated with actinomycin D for the indicated times; total RNA was prepared and analyzed by Northern blotting using a probe to the firefly luciferase open reading frame. Rp49 denotes the mRNA encoding ribosomal protein L32, a highly stable mRNA, which served as an internal loading control. (C) Stability of hid reporter mRNAs containing wild type (WT) or mutant (MT) MREs. All procedures were exactly as in (B).
Figure 1—figure supplement 1. Sequences of 3’ UTRs of reaper and hid with wild type and mutant MREs highlighted.

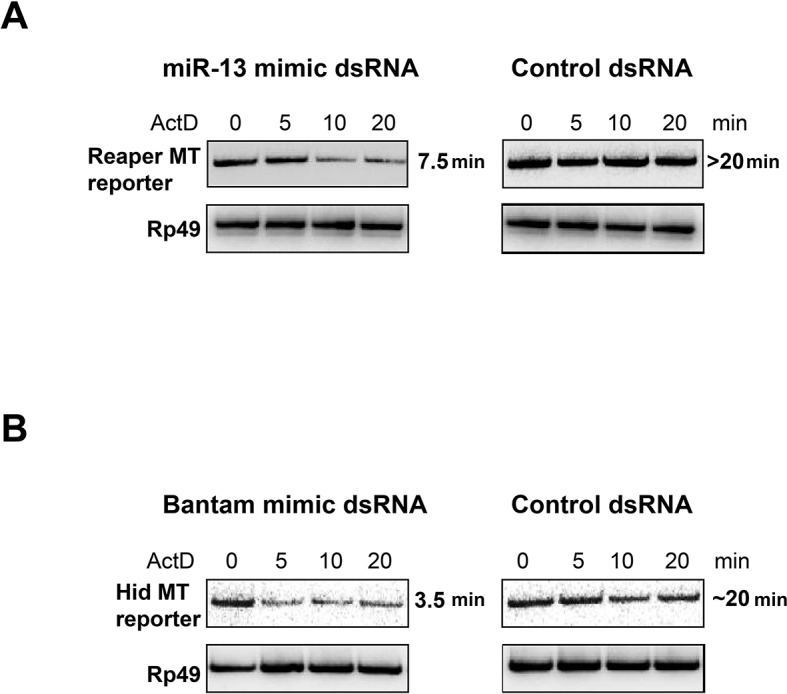
Figure 1—figure supplement 2. miRNA mimics complementary to mutant MREs destabilize reaper and hid reporter mRNAs.