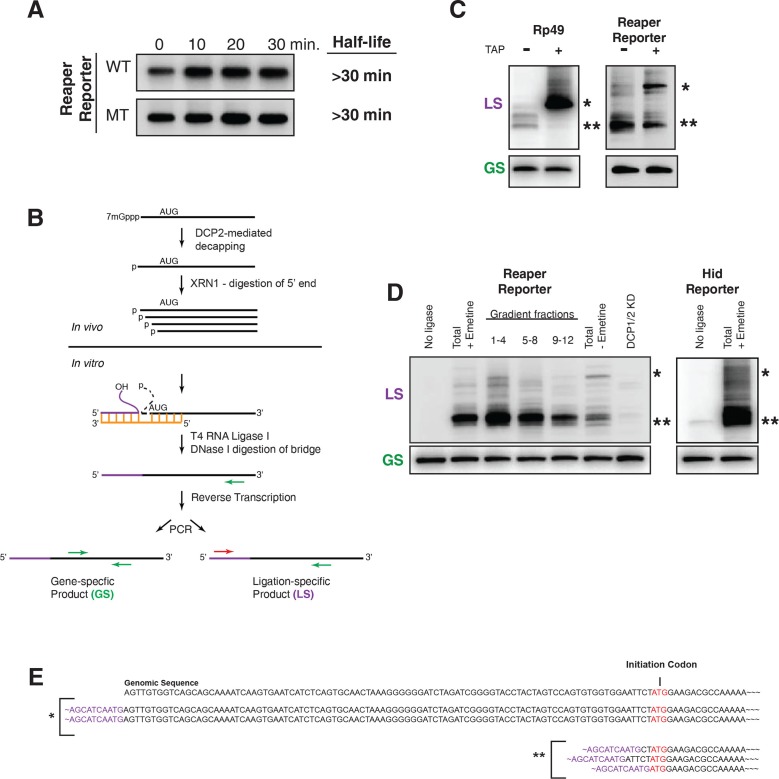
Figure 5. Cotranslational miRNA-mediated decay involves mRNA decapping.
(A) Stabilization of reaper reporter mRNAs upon knock down of Drosophila Dcp1 and Dcp2. Northern blot analysis of total RNA prepared at the indicated time after addition of actinomycin D and probed with a fragment that hybridized to the 3’ region of the firefly luciferase open reading frame. (B) Schematic depiction of the ligation strategy to identify decapped 5’ phosphorylated mRNA fragments. (C) Ligation assay for the indicated mRNAs either untreated or treated with tobacco acid pyrophosphatase (TAP). Ligation specific (LS) (decapped RNA) or gene specific (GS) PCR fragments; see (B), Heterogeneity results from digestion by XRN1 following decapping. The single asterisk denotes the position of the product when ligation is at the 5’ terminus of the mRNA. The double asterisk denotes ligation products near the AUG initiation codon. (D) Ligations assays on RNA samples from cytoplasmic extract and gradient fractions, or total RNA with or without knock down of decapping enzymes. Labelling is as in (C). (E) Sequencing analysis of the products when ligation is to the 5’ terminus of the mRNA and near the ATG initiation codon. Ligation-specific forward primer is shown in purple.