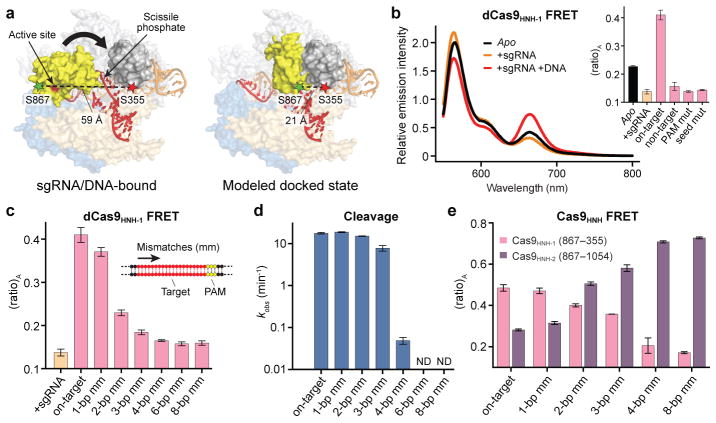
Figure 2. FRET experiments reveal an activated conformation of the HNH nuclease domain.
a, Design of Cas9HNH-1 FRET construct. Measured distances between S355 and S867 in the sgRNA/DNA-bound Cas9 structure16 and a model of the HNH domain docked at the cleavage site are indicated, as are putative conformational changes of the HNH domain (arrow). The model was generated using an HNH homolog structure (PDB ID 2QNC, ref. 21). b, Fluorescence emission spectra for dCas9HNH-1 in the presence of the indicated substrates. Inset: (ratio)A values; mut, mutation. c, (ratio)A data for dCas9HNH-1. Mismatches were introduced sequentially from the PAM-distal end of the target. d, Cleavage rate constants using wild-type Cas9. ND, cleavage not detected. e, (ratio)A data for catalytically active Cas9HNH-1 and Cas9HNH-2. Error bars in b–e represent the standard deviation; n=3.