Fig. 1.
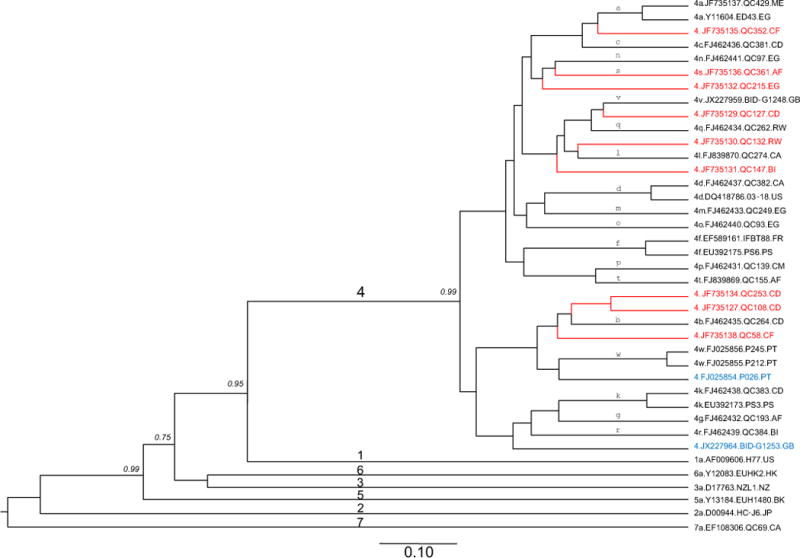
MCC tree estimated using full-length genome sequences of HCV. Reference sequences from the confirmed subtypes of genotypes 1, 2, 3, 4, 5, 6, and 7 are included (black tips), together with the nine new genomes determined in this study (red tips) and two additional unclassified variants (blue tips). Each genotype is denoted with a single digit number on the major branches of the tree while all subtypes of genotype 4 are labeled with single lower case letters above the related branches in proximity to the right tips. Isolates are named using the following format: genotype or subtype.accession number.isolate name.sampling country. To simplify the tree, only the clade posterior probability supports of less than 1 are shown in italics, otherwise they are equal to 1 but are not shown. A scale bar at the bottom represents 0.10 units of nucleotide substitution per site. Country codes: BI: Burundi; CA: Canada; CD: Congo; CF: Central Africa; CM: Cameroon; EG: Egypt; FR: France; GB: United Kingdom; HK: Hong Kong; JP: Japan, ME, Montenegro; NZ: New Zealand; PT: Portugal; RW: Rwanda; US: the United States.
