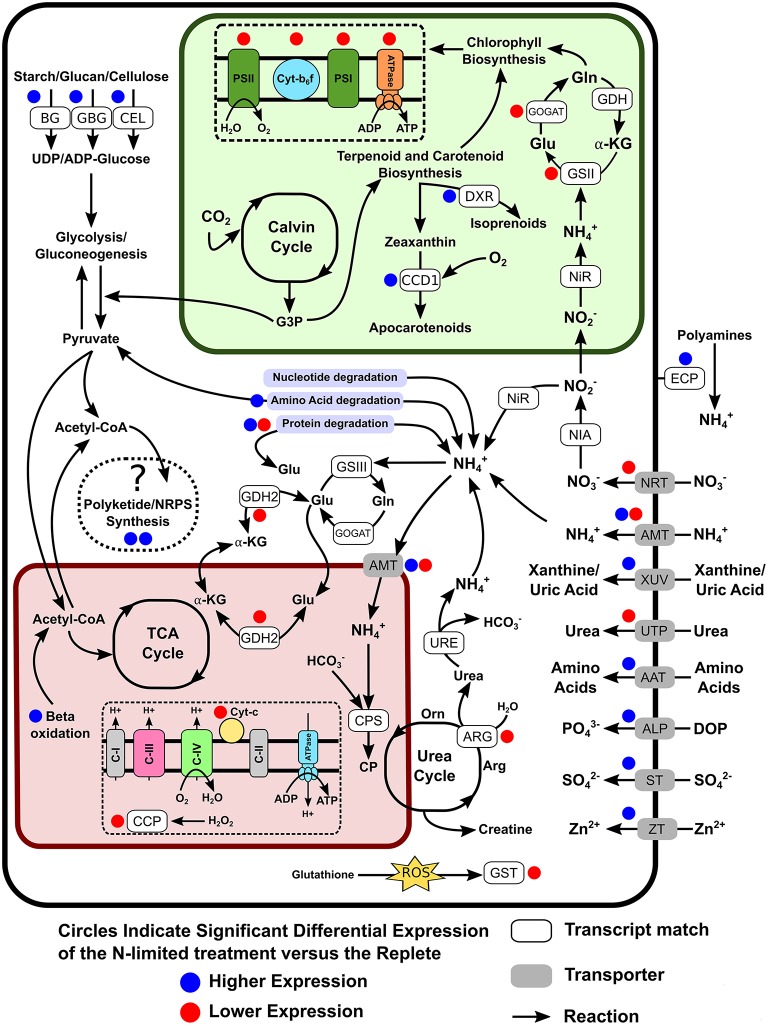
Figure 5.
Cellular overview of genes detected as differentially expressed and their potential roles in altering the metabolic pathways of S. trochoidea under N-limitation relative to replete conditions. The glyoxylate cycle is not shown, connecting the mitochondrial and plastid pathways for clarity. Blue and Red circles indicate up- and down regulation in response to N-limitation respectively. AAT, Amino acid transporters; ALP, Alkaline phosphatase; AMT, Ammonium transporter; ARG, Arginase; BG, Beta-glucanase and 1,4-beta-D-glucan cellobiohydrolase; CCP, Cytochrome c peroxidase; CCD1, Carotenoid 9, 10(9′,10′)-cleavage dioxygenase 1; CEL, Cellulase/Endoglucanase 5 precursor; CP, Carbamoyl phosphate synthetase; DOP, Dissolved organic phosphate; DXR, 1-deoxy-D-xylulose 5-phosphate reductoisomerase; ECP, Extracellular peptidases/protease; GBG, Glucan 1,3-beta-glucosidase; GDH2, Glutamate dehydrogenase 2; GSII, Glutamine synthetase II; GSIII, Glutamine synthetase III; GST, Glutathione S-transferase; GOGAT, Glutamate synthetase; NIA, Nitrate reductase; NiR, Nitrite reductase; NRT, Nitrate/Nitrite transporter; STP, Sulfate transporter; URE, Urease; UTP, Urea transporter; XUV, Xanthine/Uric Acid transporter; ZT, Zinc transporter.