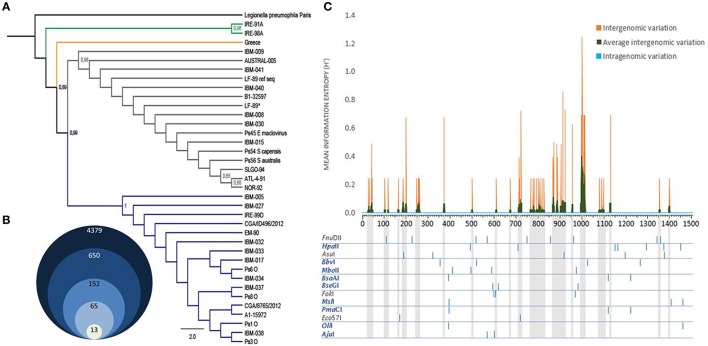
Figure 1.
P. salmonis 16S rDNA gene characterization. (A) Maximum-likelihood tree resulting from the analysis of 16S rDNA sequences of P. salmonis. Thirty six filtered sequences of 1251 bp of alignment length were used as ingroup. Legionella pneumophila strain Paris 16S rDNA sequence was used as outgroup (black branch). The numbers along the branches indicate bootstrap support values (only values ≤ 0.65 are shown). Green branches correspond to two Irish strains and the orange branch to one Greek strain. LF-89 ref seq represents the strain's 16S rDNA reference sequence in NCBI. LF-89* represents the strain's 16S rDNA sequenced in this study (Supplementary Table S1). Blue and gray branches represent two genetically distant groups formed by the other 33 strains, including all Chilean native strains (Supplementary Table S1). (B) Grouped diagram showing the initial number of restriction enzymes and the selected ones used for final analyses (for restriction enzymes selection criteria see Section Materials and Methods). (C) Intergenomic variation rate was measured as the Shannon information entropy at each position of P. salmonis 16S rDNA (orange) and subsequently averaged by a 10-bp window (green). Similarly, the intragenomic variation was calculated at each position in all the copies of the complete genomes of P. salmonis. Specific restriction enzymes patterns of P. salmonis 16S rDNA for the 13 final selected enzymes are shown. Blue enzymes, enzymes with no cuts in variable regions (green peaks and gray projections).