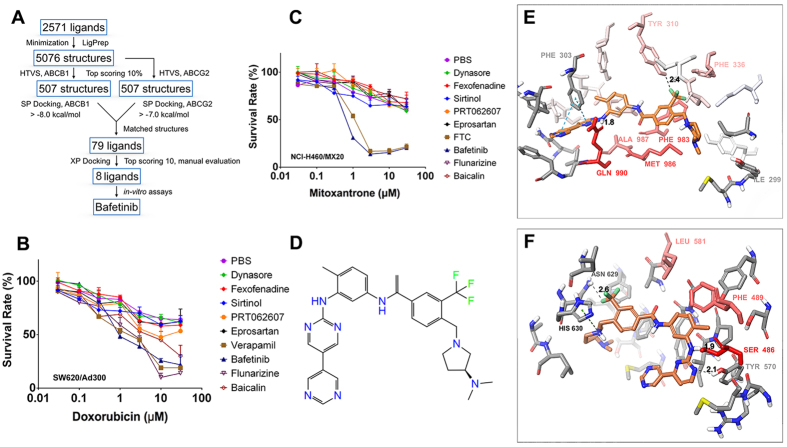
Figure 1.
(A)Schematic view of virtual screening workflow on human ABCB1 and human ABCG2. (B) The sensitization effects of virtual screened compounds on SW620/Ad300 towards doxorubicin. (C) The sensitization effects of virtual screened compounds on NCI-H460/MX20 cells towards mitoxantrone. (D) Chemical structure of bafetinib. (E) Binding geometry of bafetinib into the ABCB1 binding pocket by the Glide docking algorithms. Residues of the binding pocket are highlighted in red according to their per-residue interaction scores. Residue labels are omitted for better view except important residues suggested by per-residue interaction or residues contributed in polar interactions. Carbon atoms of bafetinib are highlighted in orange. Values of the relevant distances are given in Å. (F) Binding geometry of bafetinib into ABCG2 binding pocket. Color scheme is same as that in panel (E).