Abstract
In this issue of Cancer Cell, Pei et al. and Kawauchi et al. describe murine models of an aggressive medulloblastoma subtype driven by Myc. These tumors have a cellular origin, microscopic appearance, and molecular profile distinct from those of three other major subgroups. Thus, the models fill a significant clinical need.
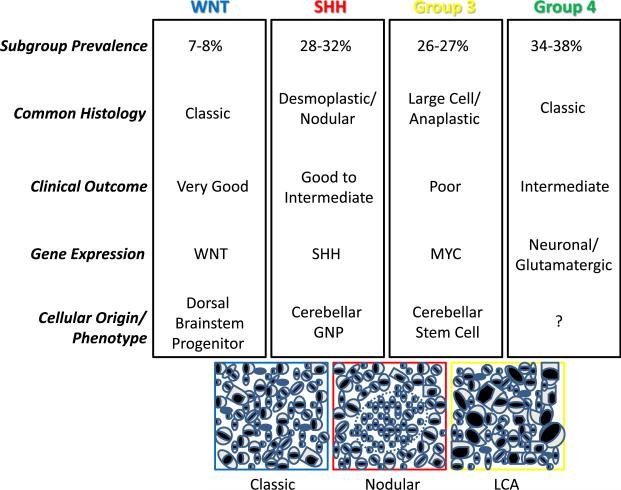
Medulloblastoma are embryonal tumors involving the cerebellum and comprised of tightly packed stem-like cells that retain the capacity to differentiate along multiple lineages. Current treatments cure only a subset of patients and result in significant long-term morbidity. Thus, improved prognostic markers and therapies are clearly needed. Recent gene expression studies have divided medulloblastoma into four major molecular subtypes, many of which also have unique clinical and histopathological features (Cho et al., 2011; Northcott et al., 2011). An international consensus panel has proposed names for the four groups (Taylor et al., 2011; Figure 1). It is hoped that our improving molecular understanding of medulloblastoma will lead to more targeted therapeutic approaches, promoting reduced morbidity in children harboring less aggressive tumors and longer survival in those with more aggressive variants.
Figure 1. Medulloblastoma Subgroups.
Transcriptional profiling supports the existence of four main medulloblastoma subgroups that differ with respect to their common microscopic appearance and clinical associations. The cartoons below depict a classic medulloblastoma comprised of heterogenous embryonal cells, a desmoplastic/nodular tumor with a central region of neuronal differentiation, and a large cell anaplastic medulloblastoma with increased nuclear size and pronounced cellular molding/engulfment.
The first two medulloblastoma subgroups are defined molecularly by WNT and Sonic Hedgehog (SHH) pathway activation, respectively, with the former associated with very good clinical behavior and the latter showing fairly good outcomes in infants and intermediate ones in older individuals. Because of their roles in Turcot and Gorlin syndromes, these pathways have long been associated with medulloblastoma formation. A transgenic mouse model of WNT-induced medulloblastoma has recently been reported, with tumors arising from progenitor cells in the embryonic dorsal brainstem and lower rhombic lip but not from granule neuron precursors (GNPs) that generate most of the cells in the cerebellum (Gibson et al., 2010). In contrast, using several transgenic medulloblastoma models driven by increased Hedgehog signaling, it has been shown that even when SHH tumors are initiated in cerebellar stem cells in vivo, they must first commit to a GNP lineage before growing into neoplastic masses in the brain (Schüller et al., 2008; Yang et al., 2008).
What has been lacking until now is a well-validated model of Group 3 medulloblastoma. This is clinically important, as this subgroup is the most aggressive in terms of its growth, dissemination, and resistance to current therapies. Group 3 medulloblastoma are characterized by amplification and overexpression of the c-myc (MYC) oncogene, and many of them are of the large cell/anaplastic (LCA) histopathological subtype (Taylor et al., 2011). It has been shown that Mycn can drive medulloblastoma formation in mice, but only a subset of these show an LCA phenotype, and most Group 3 tumors are not associated with elevated MYCN levels (Swartling et al., 2010). In human medulloblastoma cell lines, the introduction of MYC can promote aggressive xenograft growth and an LCA appearance (Stearns et al., 2006). However, it has not been clear if MYC plays a role in the initiation of medulloblastoma, or what cells might be susceptible to transformation by this oncogene. The models described in this issue of Cancer Cell significantly improve our understanding of how Group 3 medulloblastoma form, and will be critical for testing potential therapies for the group of children with highly aggressive medulloblastoma, which needs them most.
Pei et al. (2012 [in this issue of Cancer Cell]) isolated cells from postnatal murine cerebellum based on expression of the stem cell marker Prominin1 (Prom1) and lack of lineage marker expression that defines GNPs. They have previously shown that this population has functional stem cell properties and resides predominantly in the cerebellar white matter. The introduction of a mutant, stabilized Myc construct was sufficient to promote proliferation and self-renewal in vitro; however, when injected into the cerebella of immunocompromised mice, cells proliferated for a few weeks but also showed significant apoptosis and did not form tumors. By introducing both stabilized Myc and dominant negative p53 (DNp53), the investigators were able to block this apoptotic induction and tumors formed in vivo within three months of injection. Wild-type Myc could also induce tumors in conjunction with DNp53, but with reduced penetrance and increased latency.
Importantly, the murine tumors recapitulated many features of human Group 3 medulloblastoma. The cells were larger than those in SHH-driven models and showed other morphological similarities to LCA tumors, including nuclear molding and prominent cell death. Gene expression analysis revealed that they were distinct from SHH-induced murine medulloblastomas and, among human medulloblastomas, were most similar to the subgroups defined by elevated MYC (Cho et al., 2011; Northcott et al., 2011). Both immunohistochemical analysis and RNA profiling suggested that the tumors were largely undifferentiated.
Kawauchi et al. (2012 [in this issue of Cancer Cell]) also introduced Myc into cerebellar cells ex vivo, but they used Trp53 null GNPs sorted using the neuronal lineage marker Atoh1. Injection of these cells into immunocompromised mice resulted in tumors with LCA features distinct from the WNT or SHH-induced medulloblastoma previously analyzed by this group, both of which have a more classic histopathological appearance. Analysis of the Myc-driven tumor transcriptome also supported the notion that they were similar to human Group 3 medulloblastoma and distinct from prior murine models induced by WNT, SHH, or Mycn. Mycn is a target of the SHH pathway in the developing cerebellum, and medulloblastoma previously generated using Mycn by this group had a gene expression profile more similar to SHH tumors than those promoted by Myc.
Kawauchi et al. (2012) also detected increased expression of Prom1 and other stem cell factors in their Myc-driven tumors despite the fact that they were initiated in sorted populations of GNPs largely negative for these markers. Indeed, Atoh1 expression was lost in tumors, suggesting that either they had arisen from rare Atoh1 negative cells in the highly enriched starting material or that sorted GNPs were de-differentiating or otherwise silencing expression of lineage markers as part of their transformation. Pei et al. (2012) also could induce tumors from sorted GNP populations and found that the resulting medulloblastoma had lost expression of neuronal lineage markers. Moreover, both groups found that expression profiles of the Myc-driven medulloblastoma showed significant overlap with those of neural stem cells, induced pluripotent stem cells, and embryonic stem cells. Together, these suggest that Group 3 medulloblastoma either arise from neural stem cells or de-differentiate and take on a stem-like phenotype as part of their Myc-induced transformation.
Both groups also began to use their new models to evaluate potential therapies for these clinically aggressive tumors. Kawauchi et al. (2012) showed that the tumors are resistant to SHH pathway inhibitors, a significant finding as these compounds have begun to enter clinical trials, and it had been suggested that their efficacy might not be limited to the SHH medulloblastoma subgroup. Pei et al. (2012) similarly found that their tumors were insensitive to SHH pathway blockade but demonstrated that ongoing Myc expression is required to maintain tumor growth. They also used gene expression profiling to identify PI3K and mTOR signaling as potential novel therapeutic targets in Myc-driven medulloblastoma, demonstrated that small molecule inhibitors of PI3K and mTOR could slow tumor cell growth in vitro, and then validated the pro-survival effects of one compound in vivo.
To improve our treatment of cancer, we must embrace and seek to understand its complexity. Over the last two decades, analysis of the pediatric brain tumor medulloblastoma has generated numerous insights into how cancer can be meaningfully subgrouped and how oncogenic stimuli associated with developmentally important signaling pathways interact with specific populations of stem and progenitor cells to induce tumors. The two studies discussed above represent the next link in this chain, providing important new tools for preclinical testing and yielding insights into the origin and nature of an aggressive medulloblastoma subgroup associated with Myc. The new models will also be useful in addressing unexpected recent findings, such as the discovery of a strong photoreceptor expression signature in Group 3 medulloblastoma.
REFERENCES
- Cho YJ, Tsherniak A, Tamayo P, Santagata S, Ligon A, Greulich H, Berhoukim R, Amani V, Goumnerova L, Eberhart CG, et al. J. Clin. Oncol. 2011;29:1424–1430. doi: 10.1200/JCO.2010.28.5148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gibson P, Tong Y, Robinson G, Thompson MC, Currle DS, Eden C, Kranenburg TA, Hogg T, Poppleton H, Martin J, et al. Nature. 2010;468:1095–1099. doi: 10.1038/nature09587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kawauchi D, Robinson G, Uziel T, Gibson P, Rehg JE, Gao C, Finkelstein D, Qu C, Pounds S. Cancer Cell. 2012;21:168–180. doi: 10.1016/j.ccr.2011.12.023. this issue. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Northcott PA, Korshunov A, Witt H, Hielscher T, Eberhart CG, Mack S, Bouffet E, Clifford SC, Hawkins CE, French P, et al. J. Clin. Oncol. 2011;29:1408–1414. doi: 10.1200/JCO.2009.27.4324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pei Y, Moore CE, Wang J, Tewari AK, Eroshkin A, Cho YJ, Witt H, Korshunov A, Read TA, Sun JL. Cancer Cell. 2012;21:155–167. doi: 10.1016/j.ccr.2011.12.021. this issue. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schüller U, Heine VM, Mao J, Kho AT, Dillon AK, Han YG, Huillard E, Sun T, Ligon AH, Qian Y, et al. Cancer Cell. 2008;14:123–134. doi: 10.1016/j.ccr.2008.07.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stearns D, Chaudhry A, Abel TW, Burger PC, Dang CV, Eberhart CG. Cancer Res. 2006;66:673–681. doi: 10.1158/0008-5472.CAN-05-1580. [DOI] [PubMed] [Google Scholar]
- Swartling FJ, Grimmer MR, Hackett CS, Northcott PA, Fan QW, Goldenberg DD, Lau J, Masic S, Nguyen K, Yakovenko S, et al. Genes Dev. 2010;24:1059–1072. doi: 10.1101/gad.1907510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taylor MD, Northcott PA, Korshunov A, Remke M, Cho YJ, Clifford SC, Eberhart CG, Parsons DW, Rutkowski S, Gajjar A, et al. Acta Neuropathol. 2011 doi: 10.1007/s00401-011-0922-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang ZJ, Ellis T, Markant SL, Read TA, Kessler JD, Bourboulas M, Schüller U, Mac-hold R, Fishell G, Rowitch DH, et al. Cancer Cell. 2008;14:135–145. doi: 10.1016/j.ccr.2008.07.003. [DOI] [PMC free article] [PubMed] [Google Scholar]