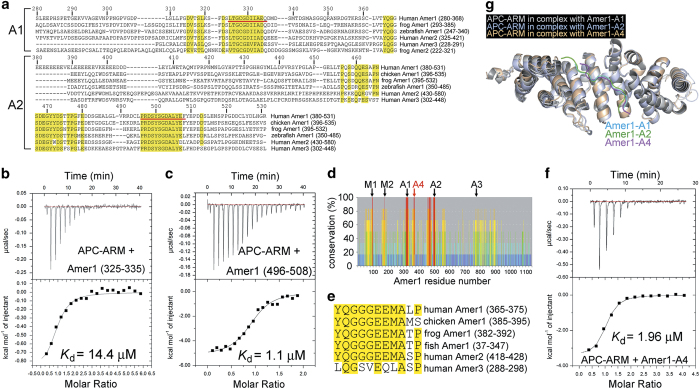
Figure 1.
Identification of a fourth APC-binding fragment A4 of Amer1/WTX, and crystal structures of APC–ARM in complex with the A1, A2, and A4 fragments of Amer1. (a) Sequence alignment of the APC-binding A1 and A2 fragments of human Amer1 (hAmer1). hAmer1-A1 (residues 280–368) and -A2 (residues 380–531) are aligned with orthologs and paralogs. The A1 and A2 fragments used in the crystallization experiments are marked with red underlines. Residues identical in all the Amer1 homologs are highlighted in yellow. Residue numbers for hAmer1 are indicated above the sequences. (b, c) The binding affinities of hAmer1 (residues 325–335, (b) and hAmer1 (residues 496–508, (c) for APC–ARM. (d) Sequence comparison of human, chicken, frog, and zebrafish Amer1, human Amer2 and Amer3 reveals a highly conserved fragment, which is named as A4. hAmer1 residue numbers are indicated, and the percentage of conservation for each residue is shown as a red, orange, yellow, green, cyan, and blue bar from high to low conservation, respectively. The membrane-binding regions M1 and M2, as well as the other APC-binding sites A1, A2, and A3, are marked. (e) Sequence alignment of the A4 site (residues 365–375) of hAmer1 and its homologs. Residues identical in at least 5 out of 6 homologs are highlighted in yellow. (f) The dissociation constant (Kd) between APC–ARM and Amer1-A4 was measured to be 1.96 μM by the ITC assay. (g) Overall crystal structures of APC–ARM in complexes with hAmer1-A1 (325–335), -A2 (496–508), and -A4 (365–375).