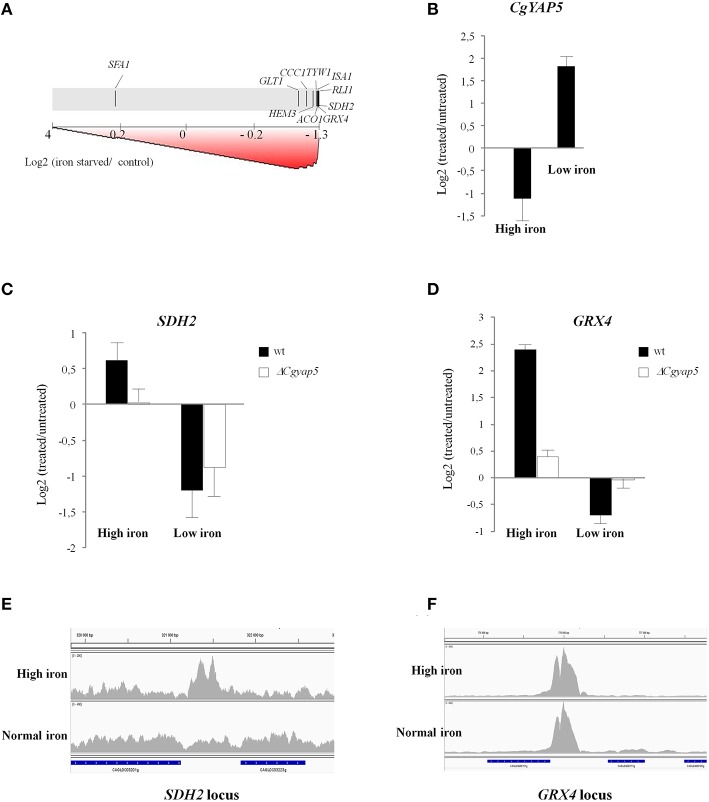
Figure 5.
CgYap5 acts as a repressor of GRX4 in iron starvation. (A) Graphical output of the Gene Set Enrichment Analyses, using as a gene set the targets of CgYap5 identified in our study and using as a test dataset the response of wild type C. glabrata cells to iron starvation caused by 0.5 mM BPS. The transcriptome data are symbolized by the gray scale with the most induced genes on the left and the most repressed on the right. The positions of the CgYap5 targets are indicated on this scale by black vertical lines. (B) The expression of CgYAP5 is inversely correlated to iron concentration. Histograms based on microarray analyses of the C. glabrata response to high iron concentration and to iron starvation caused by BPS. (C,D) SDH2 and GRX4 in response to high iron or to iron starvation, in wild type (black bars) or ΔCgyap5 (white bars) cells. The impact of the CgYAP5 deletion on the repression of GRX4 in low iron conditions was confirmed by Q-RT-PCR (Supplementary File S11). (E,F) Binding of CgYap5 to the promoters of SDH2 and GRX4 in high iron or in normal iron (YPD media) conditions (ChIP-seq).