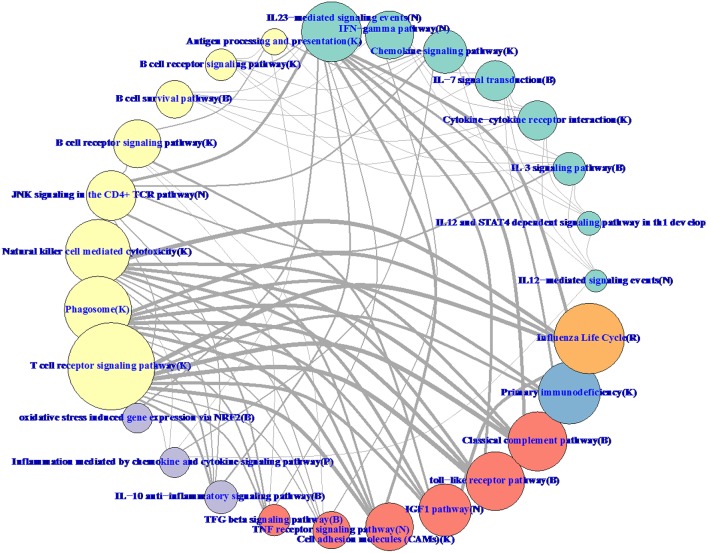
Figure 11.
RIP-M pathway-collapsed network for RNA-Seq influenza vaccine data with 4965 transcripts showing highest variation. For each of the 27 RIP-M modules, genes and their within-module edges are collapsed into one node and labeled by its top immune system pathway. The radius of each node is proportional to the number of genes in the module. The weight of each edge is the total number of edges between genes of the two modules. The top five connections are shown for each module for clarity. The modules were obtained by hard threshold τ = 0.2 of the absolute value of Pearson correlation. Colors were chosen based on pathway-type: yellow T-cell, B-cell, NK-Cell; green cytokine; purple inflammatory; blue immunodeficiency; orange influenza life cycle; red TLR and complement.