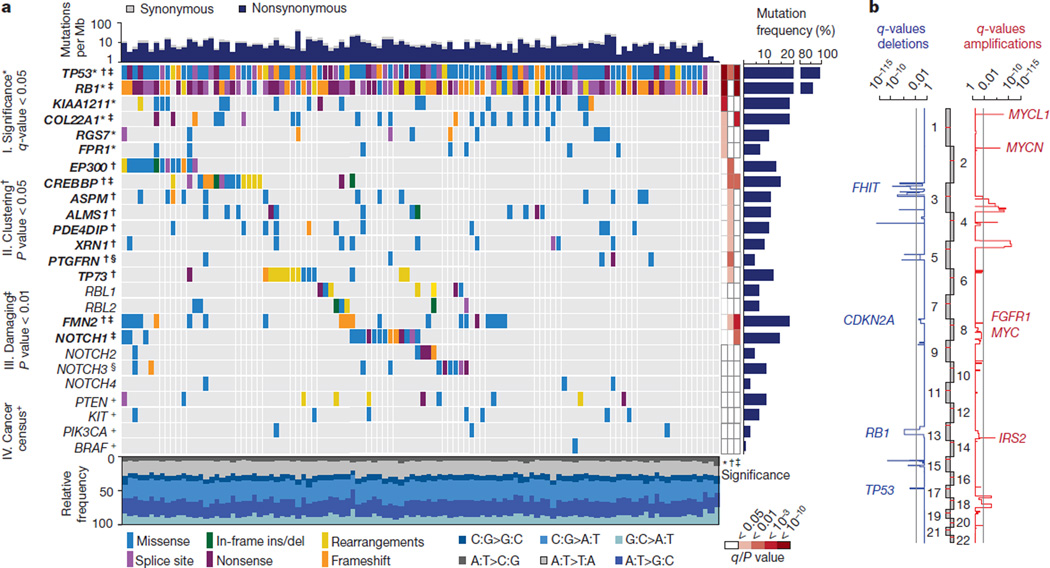
Figure 1. Genomic alterations in small cell lung cancer.
a, Tumour samples are arranged from left to right. Alterations of SCLC candidate genes are annotated for each sample according to the colour panel below the image. The somatic mutation frequencies for each candidate gene are plotted on the right panel. Mutation rates and type of base-pair substitution are displayed in the top and bottom panel, respectively. Significant candidate genes are highlighted in bold (*corrected q-values < 0.05, †P < 0.05, ‡P < 0.01). The respective level of significance is displayed as a heatmap on the right panel. Genes that are also mutated in murine SCLC tumours are denoted with a § symbol. Mutated cancer census genes of therapeutic relevance are denoted with a + symbol. b, Somatic copy number alterations determined for 142 human SCLC tumours by single nucleotide polymorphism (SNP) arrays. Significant amplifications (red) and deletions (blue) were determined for the chromosomal regions and are plotted as q-values (significance < 0.05).