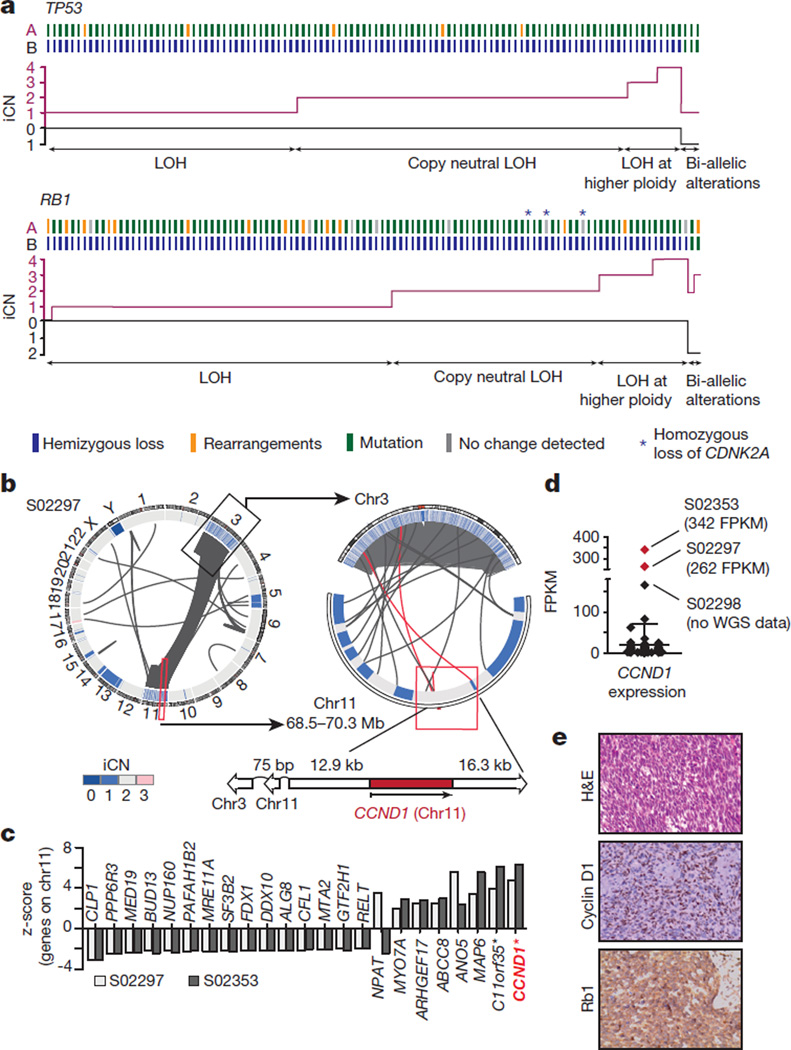
Figure 2. Universal bi-allelic inactivation of TP53 and RB1 in human SCLC.
a, Alterations of TP53 and RB1 were determined based on whole-genome sequencing data of 108 SCLC cases. Samples are plotted from left to right. Alleles A and B are represented for each case and colour-coded according to the somatic alteration. The integral copy number (iCN) state of each allele is plotted; hemizygous losses are annotated as loss of heterozygosity (LOH), copy-neutral LOH or LOH at higher ploidy. Samples retaining allele A and B show alterations on both alleles (bi-allelic alterations). b, Circos plot of case S02297 showing intra- and interchromosomal translocations between chromosome 3 and 11. The copy number state of the respective chromosomal regions (iCN) is plotted as a heatmap. The genomic context of CCND1 (on chromosome 11) is highlighted. c, Significantly differentially expressed genes encoded on chromosome 11 are analysed in both chromothripsis cases in comparison to all other tumours. Positive and negative z-scores show upregulation and downregulation of genes, respectively (P < 0.05; *q-value < 0.05). d, Distribution of CCND1 expression over 81 SCLC samples. Chromothripsis cases are highlighted in red. e, Haematoxylin and eosin (H&E) and immunohistochemistry staining for cyclin D1 and Rb1 for sample S02297. Original magnification, ×400.