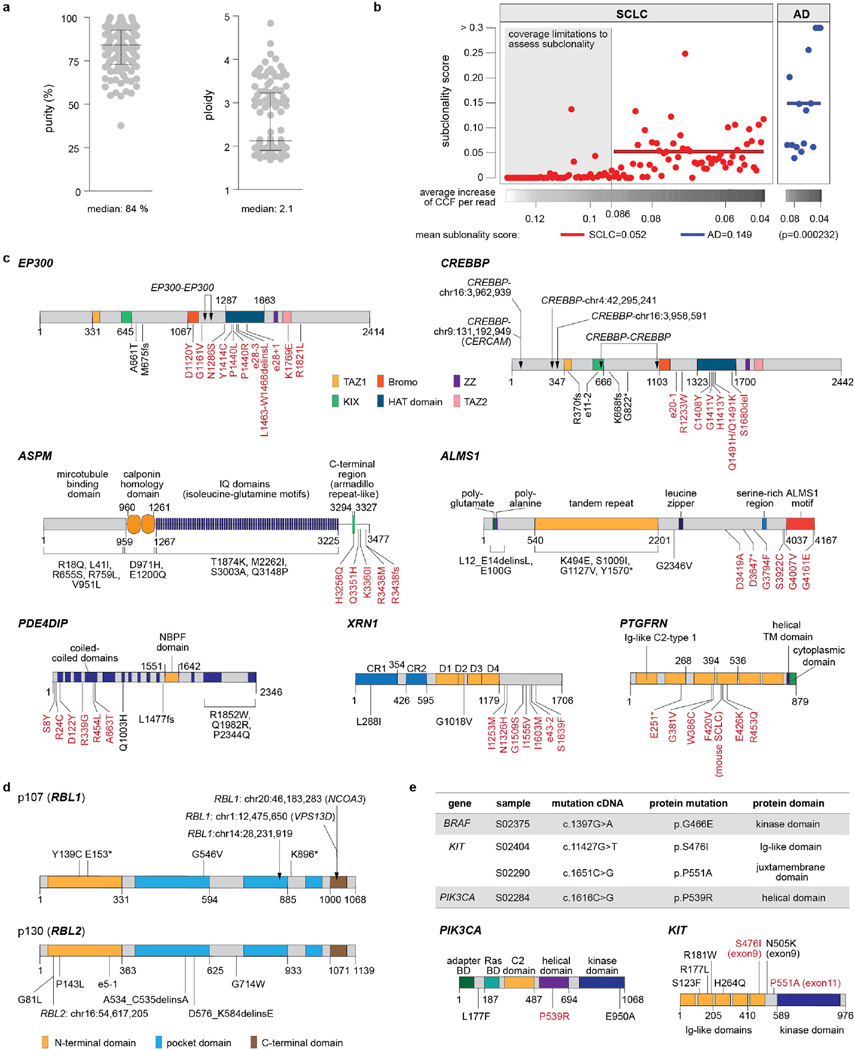
Extended Data Figure 3. Genomic characterization of SCLC tumours.
a, Purity and ploidy determined in SCLC tumours by whole-genome sequencing presented as dot density plots showing median and the interquartile range (IQR) b, Subclonal architecture of SCLC in comparison to lung adenocarcinoma (AD). Whole-genome sequencing data of SCLC and of adenocarcinoma (n = 15)11 was analysed for the presence of subclonal populations using clustering of the derived cancer cell fraction (CCF) of all single nucleotide mutations. To compare the emerging subclonal structure, we derived a subclonality score that takes into account the CCF of each subpopulation as well as its mutational burden (see Methods). In order to prevent the low sequencing coverage (35 × for SCLC and 63 × for AD) from causing a systematic underrepresentation of the subclonal diversity in the mutation calls, we computed the contribution of a single read to the CCF on genome-wide average. After systematically determining a threshold within the average increase of CCF per read values (see Methods for details), we determined the group of samples for which a reliable estimation of the subclonality score is not possible (grey area). The subclonality scores of the remaining SCLC cases were then compared to those of the adenocarcinoma cases (P = 0.000232; Mann–Whitney test). c, Schematic representation of candidate genes with significant clustering of mutations in respective protein domains. Somatic mutations and genomic translocations are mapped to the respective protein regions. Hotspot mutations are highlighted in red. d, e, Genomic alterations in the RB1 family proteins p107 (RBL1) and p130 (RBL2) (d), and in KIT and PIK3CA (e). Somatic mutations in therapeutic target genes are listed and mapped to the protein domains of KIT and PIK3CA. Mutations with potential therapeutic implications are highlighted in red.