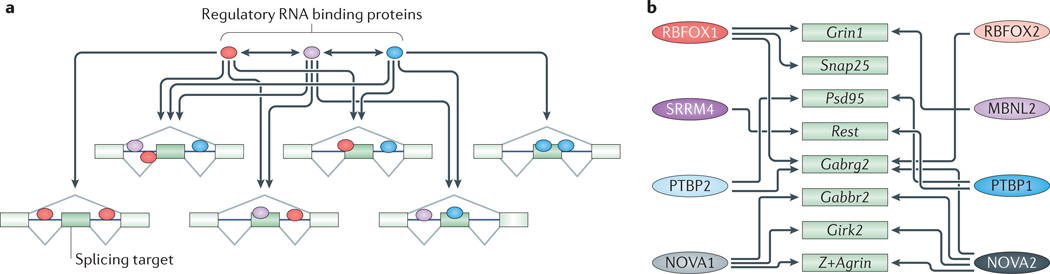
Figure 1. Splicing regulatory networks.
a | Splicing regulators control large target exon sets that often overlap with those regulated by other RNA-binding proteins (RBPs). Therefore, splicing of a given alternative exon can be affected by multiple RBPs. RBPs also affect their own splicing and homeostatic expression as well as that of other RBPs. The high degree of cross-regulation (indicated by arrows) between splicing regulators and their target sets creates complex splicing networks where the perturbation of a single RBP can lead to pleiotropic effects. Conversely, the splicing outcome of an exon can result from the combinatorial control of many RBPs. b | This figure shows some of the splicing target transcripts discussed in this Review (green boxes) that are cross-regulated (indicated by arrows) by multiple RBPs (coloured ovals on the left and right). Girk2, inwardly rectifying potassium channel Kir3.2; Gabrg2, GABAA receptor subunit gamma 2; Gabbr2, GABAB receptor 2; MBNL, muscleblind-like; NOVA, neuro-oncological ventral antigen; Psd95, postsynaptic density protein 95; PTBP, polypyrimidine tract binding protein; Rest, repressor element 1-silencing transcription factor; RBFOX, RNA-binding protein fox 1 homologue; Snap25, synaptosomal-associated protein 25; SRRM4, serine/arginine repetitive matrix protein 4.