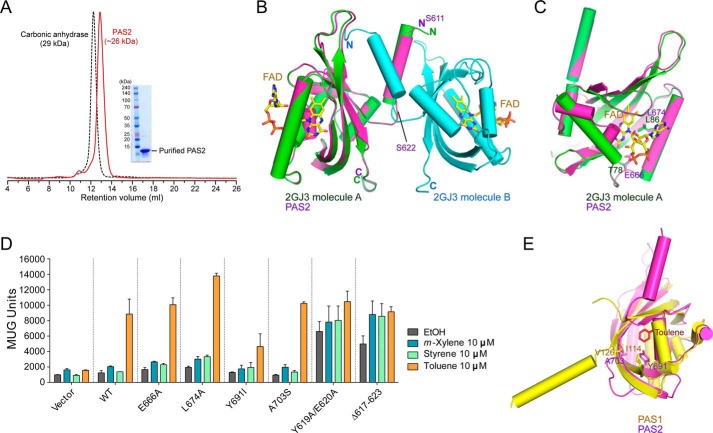
FIGURE 4.
Role of PAS2 in TodS/TodT signal transduction. A, SEC analysis of purified PAS2(611–729). Eluted PAS2 protein was compared with the molecular weight standard marker carbonic anhydrase (29 kDa) and analyzed as a dimer in solution. B, the modeled PAS2 structure (magenta) based on the dimeric structure (green and cyan) of the A. vinelandii NifL LOV domain (Protein Data Bank code 2GJ3). FAD binding to each NifL PAS domain is shown. The N-terminal α-helix (residues Ser611–Ser622) of PAS2 predicted to be involved in dimerization is indicated. C, PAS2 residues Glu666 and Leu674, corresponding to the NifL FAD binding residues Thr78 and Leu86. D, potential PAS2 residues involved in ligand binding were assessed using the β-galactosidase assay with different ligands. The N-terminal PAS2 α-helix (residues Ser611–Ser622) predicted to be involved in dimerization, as seen in Fig. 4B, was also evaluated with the same assay. E, the modeled PAS2 was superimposed onto toluene-bound PAS1. The PAS2 residues Tyr691 and Ala703 were analyzed with the corresponding PAS1 residues Ile114 and Val126, which are responsible for toluene binding.