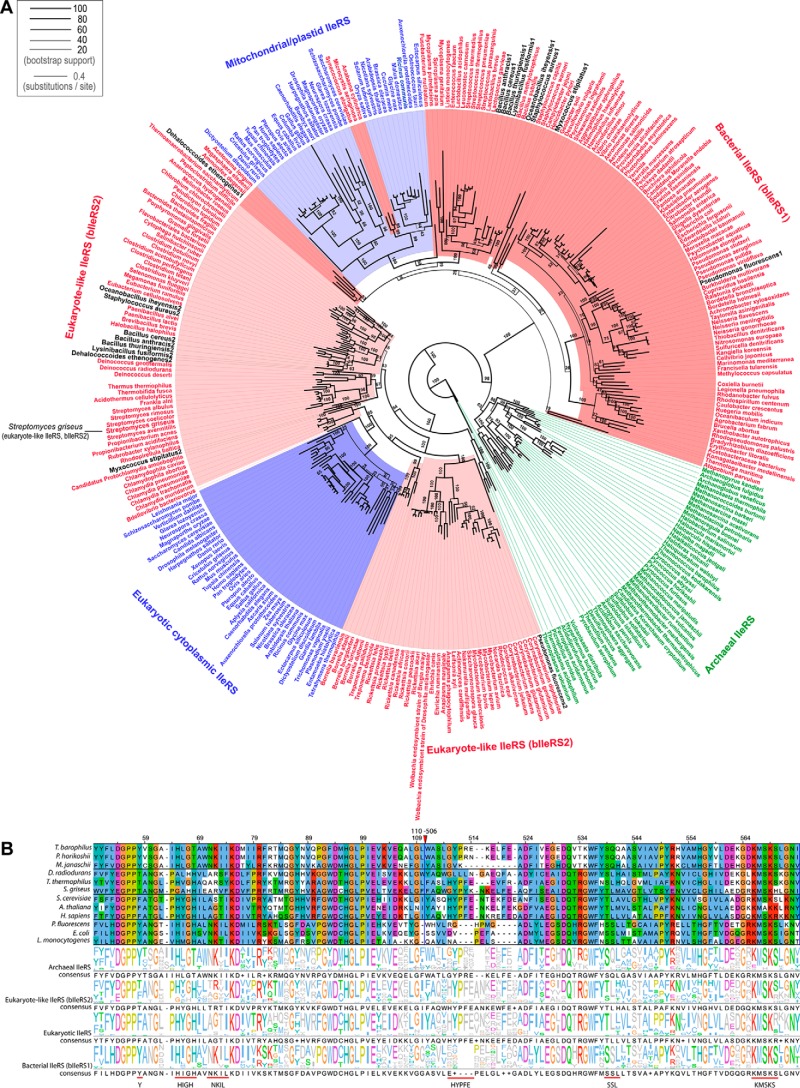
FIGURE 2.
IleRS phylogeny. A, IleRS phylogenetic tree. Bacterial species are colored red (species with eukaryote-like IleRS are shown in light red); archaea are green, and eukaryotes are blue (mitochondrial/plastid IleRS are shown in light blue). Species with both bacterial IleRS (bIleRS1) and eukaryote-like IleRS (bIleRS2) are shown in black. The line width of each branch is scaled according to the bootstrap support value (see legend), and the numbers along the branches show the percentage reproducibility of nodes in bootstrap replicates (some values were omitted for simplicity). The scale bar represents 0.4 estimated amino acid substitutions per site. B, partial amino acid sequence alignment of representative archaeal, eukaryote-like (bIleRS2), eukaryotic, and bacterial (bIleRS1) IleRSs. The presented regions include the consensus sequences HIGH and KMSKS and characteristic bacterial (Tyr-59; 71NKIL74; 572SSL, numbering according to EcIleRS) or eukaryotic/eukaryote-like motifs (541HYPFE, numbering according to ScIleRS). Each sequence logo was determined using the complete alignment with 334 sequences and represents the conservation within a certain IleRS group. The sequences were assigned to a group according to the phylogenetic analysis. The red triangle marks the omitted portion of the alignment.