FIGURE 1.
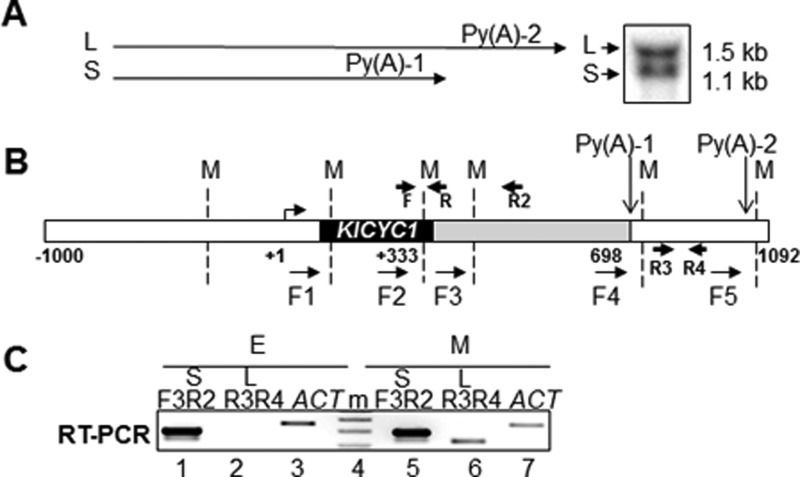
Alternative polyadenylation of K. lactis CYC1 mRNA (KlCYC1). A, schematic summary of long (L) and short (S) forms of the KlCYC1 mRNA resulting from alternative 3′-end processing at polyadenylation sites Py(A)-1 and Py(A)-2. The Northern blot depicts the L and S transcripts from cells harvested at early log phase growth (A600 = 0.6). B, schematic depiction of the KlCYC1 gene. The numbering system denotes the ATG (+1), termination codon (+333), and endonucleolytic cleavage sites as Py(A)-1 (698) and Py(A)-2 (+1092). Tandem primers used for 3C analysis are F1–F5; these primers are located adjacent to the MspI restriction sites (M) denoted by the dashed vertical lines. The primer pairs F3, R2 and R3, R4 were used to quantify KlCYC1 mRNA by RT-PCR. The F and R primers were used for quantitation of 3C chromatin. C, KlCYC1 mRNA levels as determined by RT-PCR. Lanes 3 and 7 represent mRNA encoded by the ACT1 gene and serve as loading controls. Lane 4 (m) is marker DNA used to estimate DNA length.