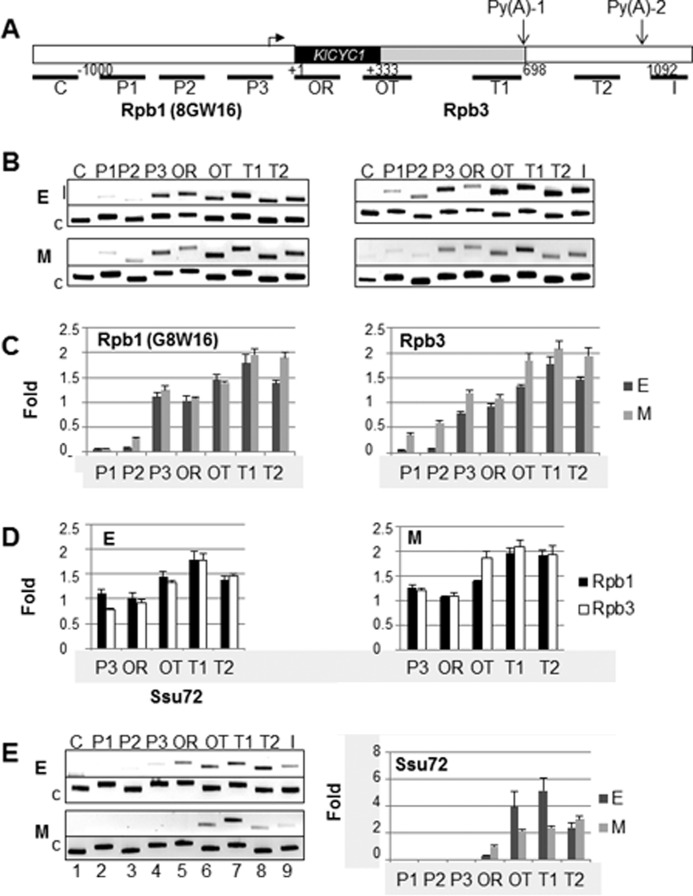
FIGURE 3.
Dependence on growth phase of RNAP II localization along the length of KlCYC1 as determined by ChIP. A, schematic indication of the KlCYC1 locus indicating the regions probed by PCR primers. P1 and P2 are upstream of the promoter; P3 encompasses the promoter; OR and OT1, probe the early ORF and immediate 3′-UTR, respectively; T1 and T2 probe DNA encoding the indicated 3′-end processing sites; I probes the region immediately downstream of the poly(A)-2 site; and C serves as a control, probing a non-transcribed region located upstream of KlCYC1. B, ChIP results for cells harvested in early log (E) and mid-log (L) phase using the 8WG16 or Rpb3 antibodies. C, quantification of Rpb1 and Rpb3 ChIP analysis comparing the growth phase (E and M). D, quantification of ChIP analysis comparing results using the Rpb1 and Rpb3 antibodies. All data are from three independent experiments equivalent to those shown in B. E, ChIP results from cells harvested in early log (E) and mid-log (M). Location of Ssu72 along KlCYC1 and its dependence on the growth phase, using the Ssu72 antibody, are shown. Error bars represent the standard error of the mean of three independent experiments.