FIGURE 5.
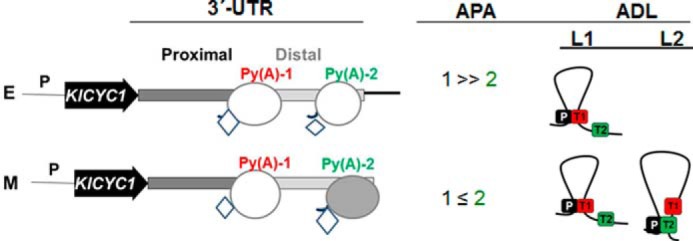
Model for the correlation between APA and alternative gene looping at the KlCYC1 locus. In early (E) log phase cells, 3′-end processing occurs predominantly at the proximal poly(A) site, which is the predominant site for looping between the promoter (P) and poly(A)-1 site (Py(A)-1) (red). Conversely, in mid-log phase (M) cells, RNAP II exhibits enhanced bypass at the proximal poly(A)-1 (Py(A)-1) (red) and processing to the poly(A)-2 site (Py(A)-2) (green), resulting in approximately equal 3′-end processing at the two poly(A) sites, coinciding with comparable looping between these two sites and the promoter. Key: RNAPII (oval) and Ssu72 (diamond) during the different phases of cell growth (E and M). ADL denotes alternative looping being L1 and L2, the two alternatives. RNAPII is in gray color when Rpb3 abundance is higher than hypophosphorylated Rpb1. The increase in diamond size reflects changes of the abundance of this factor.
