Figure 2.
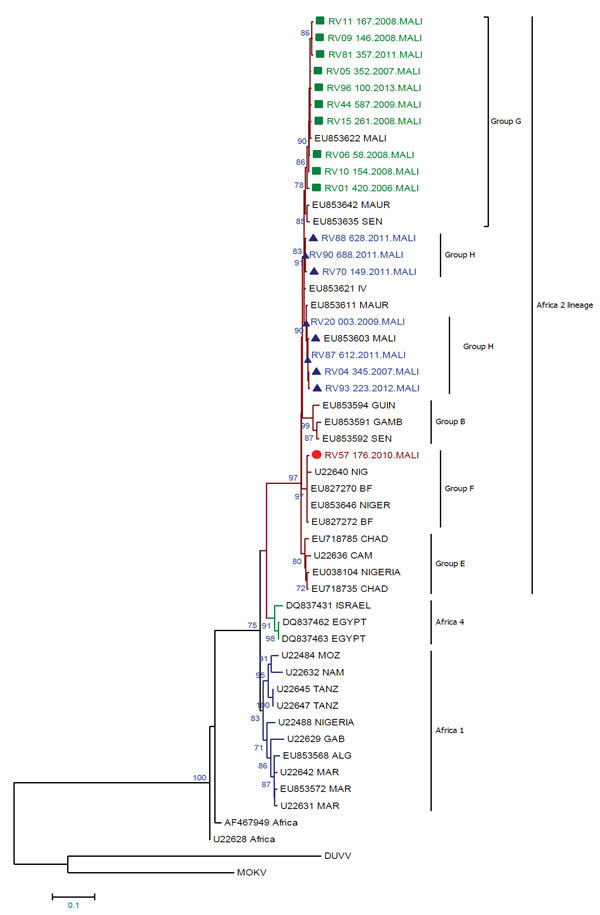
Maximum-likelihood phylogenetic tree based on a 564-nt sequence of nucleoprotein genes of 18 rabies virus sequences from Mali, 2002–2013, and representative sequences from Mali (n = 2), northern Africa (n = 6), South Africa (n = 2), West Africa (n = 32), and central Africa (n = 5). Sequences obtained in this study are identified in green, blue, and red. Green squares indicate genotype G, blue triangles indicate genotype H, and red circles indicate genotype F. The tree is rooted with 2 bat isolates used as outgroups Duvenhage virus (DUVV) (U22848) and Mokola virus (MOKV) (U22843). Bootstrap values (100 replicates) >70% are shown next to nodes. Alg, Algeria; BF, Burkina Faso; Cam, Cameroon; Gab, Gabon; Gamb, Gambia; Guin, Guinea; Maur, Mauritania; Mar, Morocco; Moz, Mozambique; Nig, Niger; Sen, Senegal; Tanz, Tanzania. Scale bar indicates nucleotide substitutions per site.
