Figure 1.
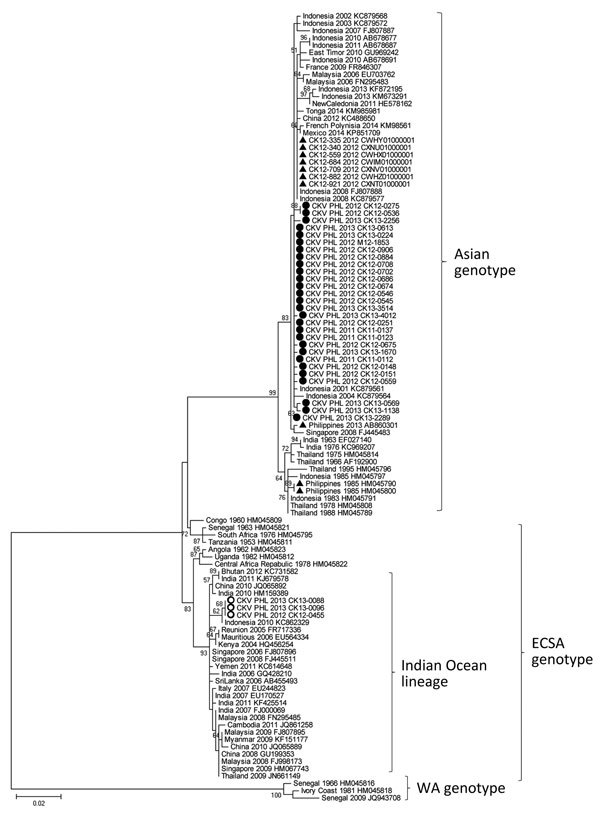
Phylogenetic analysis of partial (733 nt) E1 gene of 31 CHIKVs detected in the Philippines in this study during 2011–2013 compared with 77 global strains. The tree was constructed using maximum-likelihood method with the Kimura 2-parameter model and 1,000 bootstrap replications. Bootstrap values >50% are indicated on the branches of the tree. Black circles indicate Asian genotypes; open circles indicate ECSA genotypes analyzed in this study; triangles indicate reference strains collected in the Philippines. CHIKV, chikungunya virus; ECSA, East/Central/South African, WA, West African. Scale bar indicates nucleotide substitutions per site.
