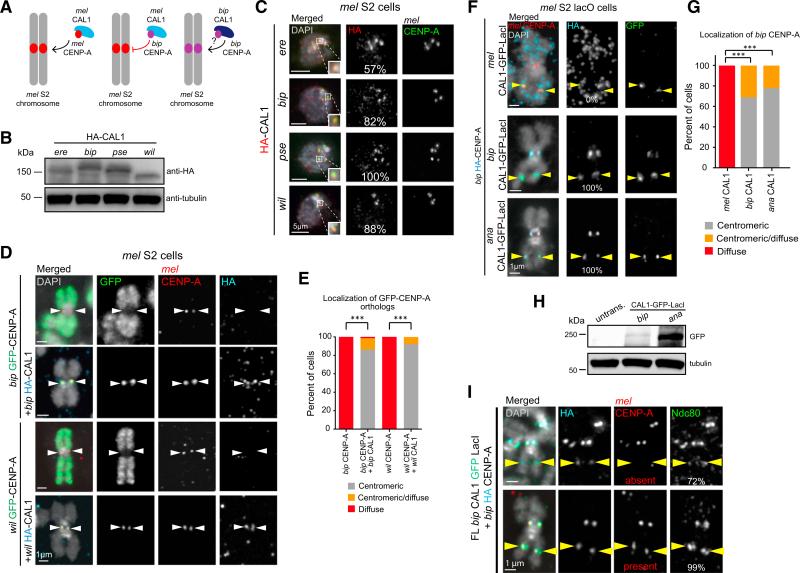
Figure 3. Co-expression of CAL1 and CENP-A Ortholog Pairs Rescues Centromeric Localization.
(A) Schematic of the experiments testing whether co-expression of bip CAL1 and bip CENP-A can result in the localization of bip CENP-A to mel centromeres.
(B) Western blots of whole-cell extracts showing expression of HA-CAL1 constructs. Top: anti-HA. Bottom: anti-tubulin (loading control).
(C) Representative IF images of interphase mel S2 cells transiently expressing HA-CAL1 orthologs. DAPI is shown in gray, HA in red, and mel CENP-A in green. The percentage of cells with centromeric HA signal is indicated in the middle column. Zoomed panels show representative centromeres with merged colors.
(D) Representative IF images of metaphase chromosome spreads from S2 cells transiently expressing bip or wil GFP-CENP-A alone (first and third rows, respectively), bip GFP-CENP-A and bip HA-CAL1 (second row), or wil GFP-CENP-A and wil HA-CAL1 (fourth row). DAPI is shown in gray, GFP in green, HA in aqua, and mel CENP-A in red. White arrowheads indicate the position of the centromere.
(E) Quantification of the IF shown in (D). Chromosome spreads were manually classified as having either centromeric (gray bars), diffuse (red), or centromeric/ diffuse (orange) GFP signal. n = 29 spreads for bip CENP-A alone, 73 for bip CENP-A with bip CAL1, 27 for wil CENP-A alone, and 51 for wil CENP-A with wil CAL1. ***p < 0.0001 (Fisher's two-tailed test) for the centromeric localization of bip and wil CENP-A with and without CAL1. These data were confirmed by one biological replicate using HA-tagged CAL1 constructs (data not shown), and two biological replicates using CAL1-GFP-LacI and HA-CENP-A constructs (see F in this figure for bip, and data not shown for wil). See also Figures S4 and S5.
(F) Representative IF images of metaphase chromosome spreads from mel lacO S2 cells transiently co-expressing mel, bip, or ana CAL1-GFP-LacI (first, second, or third row, respectively), and bip HA-CENP-A. GFP is shown in green, HA in aqua, mel CENP-A in red, and DAPI in gray. Yellow arrowheads indicate the position of the lacO site.
(G) Quantification of the images shown in (F). Cells were manually classified as having either exclusively centromeric GFP signal (gray bars), diffuse GFP signal (red bars), or centromeric and diffuse GFP signal (orange bars). n ≥ 30 cells per condition. These data were confirmed by two biological replicates (data not shown). ***p < 0.0001 (Fisher's two-tailed test).
(H) Western blots with anti-GFP (top) and anti-tubulin (loading control, bottom) antibodies of whole-cell extracts showing the expression of induced bip and ana CAL1-GFP-LacI in lacO cells in F.
(I) Representative IF images of metaphase chromosome spreads from mel lacO S2 cells transiently co-expressing bip CAL1-GFP-LacI and bip HA-CENP-A (aqua) showing the lacO recruitment of endogenous mel CENP-A (red) and the outer kinetochore protein Ndc80 (green). GFP fluorescence was quenched with 100% ethanol. DAPI is shown in gray. Percentage of CENP-A positive (bip, or mel and bip) lacO arrays with Ndc80 is indicated in the right column. Yellow arrowheads mark the position of lacO site. These data represent the average of three experiments (two technical replicates and one biological replicate). n = 40 HA-positive cells.