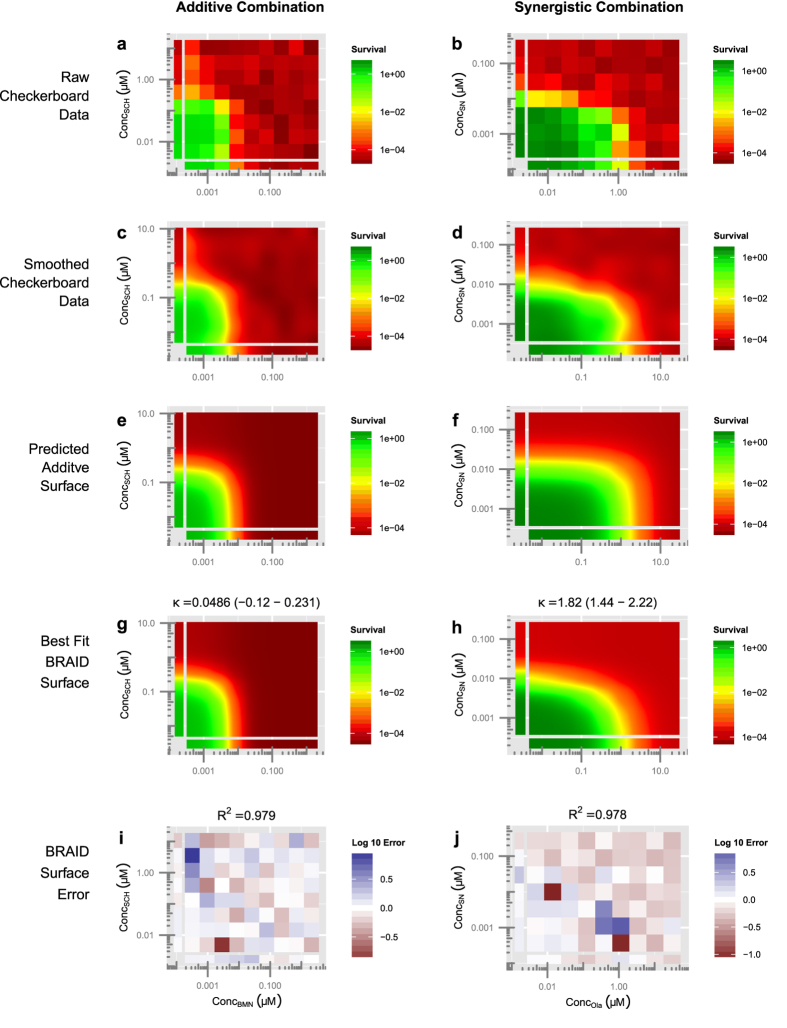
Figure 4. Illustration of the BRAID analysis approach.
The combination of BMN-673 and SCH-900776 in cell line ES8 (a,c,e,g,i) and the combination of olaparib and SN-38 in cell line ES1 (b,d,f,h,j). (a,b) Cell survival (modeled by Cell-Titer Glo luminescence) is calculated relative to negative controls on all plates, and the average logarithm of each dose-pair is plotted as a response surface. (c,d) Smoothing data using a simple Gaussian interpolation allows the viewer to quickly evaluate the shape of the response surface. (e,f) The additive BRAID surface estimated using the best-fit individual dose-response parameters of the two drugs. (g,h) The best fit BRAID surface in which the interaction parameter κ is allowed to vary freely. The best fit BRAID surface for BMN-673 vs. SCH-900776 is very close to the estimated additive surface, while for olaparib vs. SN-38 it is visibly different from the additive surface. This is due to strong synergy between olaparib and SN-38. (i,j) Comparing the best fit BRAID surface with the measured unsmoothed data allows the viewer to quickly gauge whether the surface contains non-uniform errors, suggesting artifacts or a poor fit.