Figure 9. Binding of L. lactis BirA1 to the bioY promoter region.
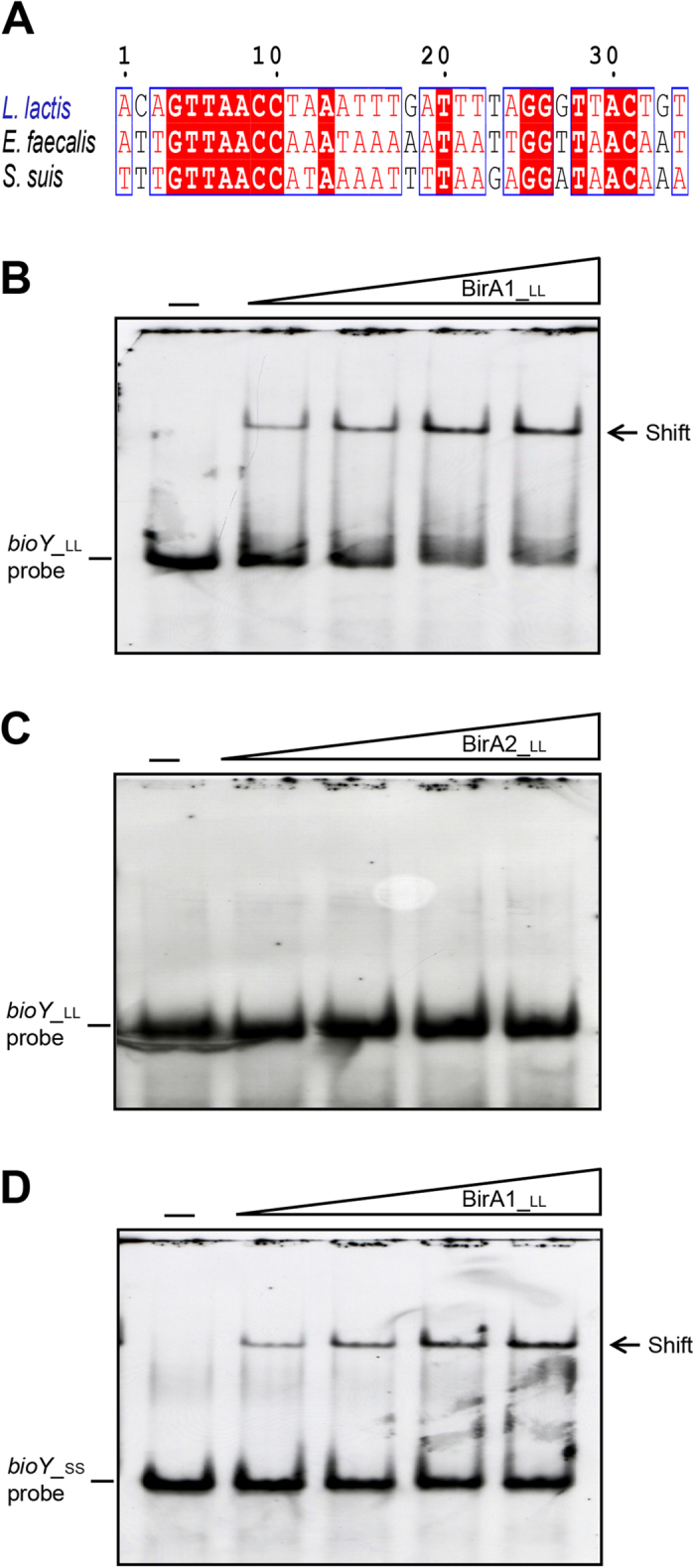
(A) Multiple sequence alignment of the predicted BirA-recognized palindromes localized in the bioY promoter regions Identical residues are in white letters with red background, similar residues are in red letters with white background, whereas non-conserved residues are in grey letters. Electrophoretic mobility shift assay (EMSA) of the binding of L. lactis BirA1 to the promoter regions of bioY from L. lactis (in Panel B) and the closely-related organism Streptococcus suis (in Panel D). (C) EMSA-based evidence that L. lactis BirA2 protein cannot bind to the promoter region of L. lactis bioY. The minus sign denotes no addition of either BirA1_LL or BirA2_LL protein, and the DIG-labeled probe shifted by BirA1_LL protein is indicated with an arrow. The concentrations of BirA1_LL (BirA2_LL) in the right four lanes of each panel were (from left to right) 3, 6, 9 and 12 pmol, respectively. The protein samples were incubated with 0.2 pmol of DIG-labeled probe in a total volume of 15 μl. To make biotinoyl-AMP, the ligand of BirA protein, 100 uM biotin plus 100 uM ATP was added into the gel shift system and incubated for ~1 hour. Designations LL and SS denote L. lactis and S. suis, respectively. BirA1_LL and BirA2_LL denote the two putative versions of L. lactis BirA protein, whereas bioY_LL and bioY_SS denote the bioY gene from L. lactis and S. suis, respectively. A representative graph from three independent gel shift assays is presented here. 7.5% native PAGE gels were used for the assays.
