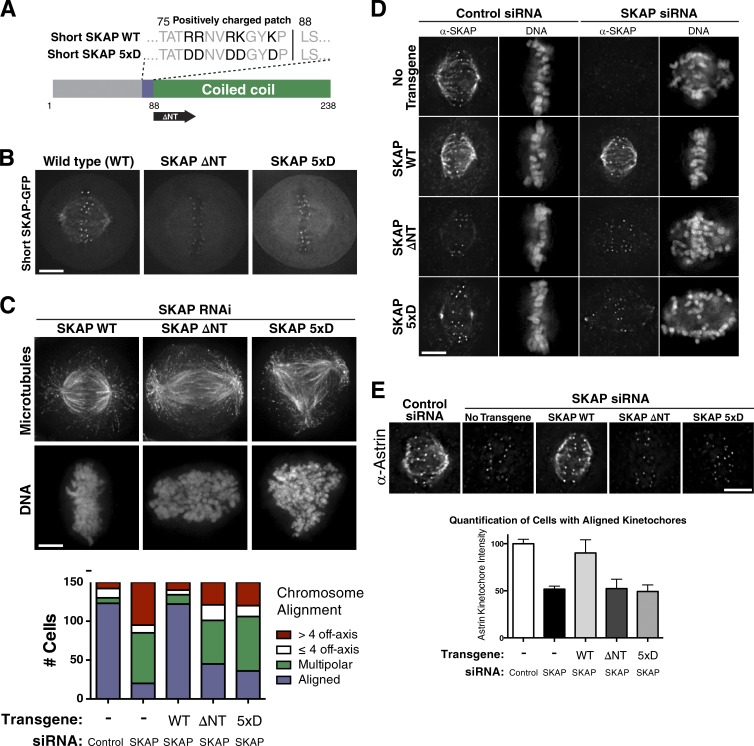
Figure 3.
SKAP microtubule-binding mutants prevent Astrin/SKAP complex microtubule localization and display defects similar to SKAP depletion. (A) Diagram of SKAP microtubule-binding mutants: a mutant with five positive charged amino acids changed to aspartate (SKAP 5xD) and mutant removing the N terminus to leave only the SKAP coiled coil (SKAP ΔNT). (B) Fluorescence images showing SKAP-GFP localization in cells expressing wild-type (WT) SKAP or the indicated SKAP microtubule-binding mutants. (C, top) IF images showing examples of cells with aligned chromosomes, many off-axis chromosomes, and a multipolar spindle from RNAi-based replacement experiments with SKAP WT, ΔNT, and 5xD, respectively. Images represent maximum-intensity projections. (bottom) Quantification of SKAP mutant cell phenotypes. (D) IF images showing SKAP and DNA localization in the indicated SKAP microtubule-binding mutants. Although most mutant cells display defective chromosome alignment, see Fig. S2 H for cells with aligned chromosomes to highlight the ability of SKAP to localize to kinetochores but not microtubules. DNA is scaled independently for each image. (E, top) IF images showing Astrin localization in the indicated conditions. See Fig. S2 I for cells displaying extreme phenotypes and misaligned plates. (bottom) Kinetochore intensity quantification for Astrin IF in indicated conditions. Each condition represents data from five cells and 50 total kinetochores. SEM is plotted for the averages from each cell in each condition. Using an unpaired two-tailed t test, control and rescue cell kinetochore intensities were statistically indistinguishable (P = 0.2464), and each was significantly different from SKAP-depleted or microtubule-binding mutants (P < 0.0001 for each). The SKAP-depleted and microtubule-binding mutant conditions were statistically indistinguishable from each other (SKAP depleted: ΔNT, P = .9110; SKAP depleted: 5xD, P = .5023; ΔNT: 5xD, P = .6038). Bars, 5 µm.