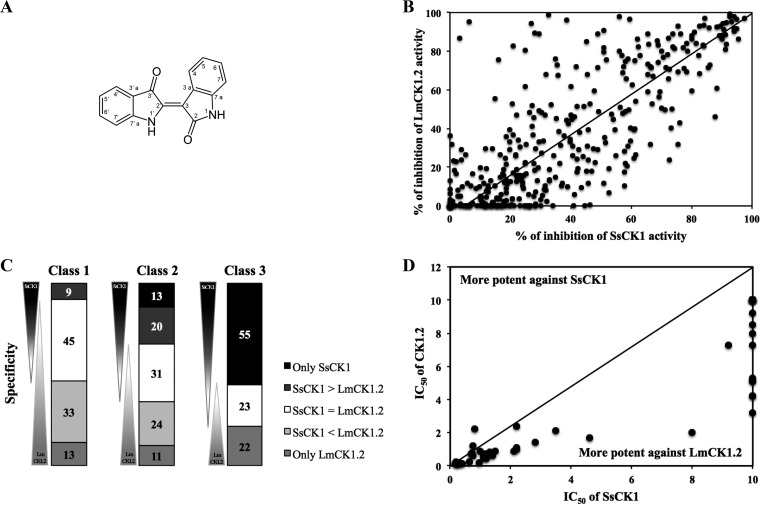
FIG 4.
Screening of the indirubin derivative library. (A) Structure of the indirubin backbone. (B) Target-based screening of 400 derivatives. Each point represents the percent inhibition toward LmCK1.2 activity versus the percent inhibition toward SsCK1 activity for each compound. The compounds in the top left are more potent toward LmCK1.2, whereas the compounds in the bottom right are more potent toward SsCK1. (C) Compounds were classified according to their specificity: only potent on SsCK1, more potent on SsCK1 than LmCK1.2 (SsCK1 > LmaCK1.2), equally on both kinases (SsCK1 = LmCK1.2), more potent on LmCK1.2 than SsCK1 (SsCK1 < LmaCK1.2), and only potent on LmCK1.2. Compounds were also classified according to their percent inhibition: class 1 corresponds to compounds that inhibit the kinases between 80 and 100%, class 2 corresponds to compounds that inhibit the kinases between 60 and 80%, and class 3 corresponds to compounds that inhibit the kinases between 40 and 60%. Fifty-seven percent of the compounds are more potent toward LmCK1.2 than SsCK1, and 46% are specific to LmCK1.2. (D) IC50s of the 55 compounds that are specific to LmCK1.2 or that belong to class 1 with a percent inhibition above 90%. Each point represents the IC50 of a particular compound toward LmCK1.2 versus SsCK1. The IC50 are lower against LmCK1.2 than SsCK1.