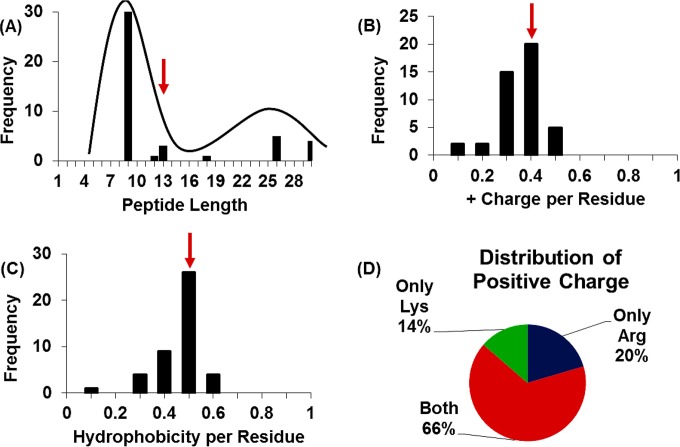
FIG 1.
Database filtering. (A) Peptide length of the database peptides shows a bimodal distribution centered around 9 and 25 residues. The selected length of the designed AMPs is 13 residues (red arrow), the mean of the entire distribution. (B) Distribution of positive charge per residue length of database peptides. A +0.4 charge per residue, the distribution mean, is selected for the designed AMPs (red arrow). (C) Distribution of the number of hydrophobic residues (Ala, Val, Ile, Leu, Trp, Phe, Met, and Cys) per total peptide length, where 0.5, the mean of the distribution (red arrow), is chosen for the designed AMPs. (D) Distribution of positive charge within database peptides. Histidine was excluded due to the limited number of peptides containing histidine in the database.