TABLE 1.
Drug killing rates for the continuous-time and discrete-time modelsa
| Drug | Half-life | Stage specificity | Continuous-time model | Discrete-time model |
|---|---|---|---|---|
| Hypothetical drug 1 | Long | No | Vmax = ln(PRR48)/48 + a | V′max = ln(PRR48)/48 + a |
| Partner drug | Long | Yes | V̂max = ln(PRR48)/48 + a | V̂′max = V̂max 48/q |
| Hypothetical drug 2 | Short | No | Ṽmax = [ln(PRR48) + 48a]/ta | Ṽ′max = [ln(PRR48) + 48a]/ta |
| Artemisinin derivative PRR48 calibration | Short | Yes |
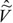 max,48 = [ln(PRR48) + 48a]/ta max,48 = [ln(PRR48) + 48a]/ta
|
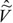 ′max,48 = ′max,48 = 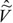 max,48 48/q max,48 48/q
|
| Artemisinin derivative PRR96 calibration | Short | Yes |
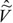 max,96 = [ln(PRR48) + 96a]/3ta max,96 = [ln(PRR48) + 96a]/3ta
|
Obtained by iteration |
a is the instantaneous parasite growth rate over the 48-h parasite RBC cycle, PRR48/PRR96 is the reduction in parasite number over 48 or 96 h (i.e., one or two parasite RBC cycles) following drug treatment (the value is different for each drug but identical for both models when used for the same drug), q is the number of 1-h bins during which killing occurs, and ta is the duration of drug action after each dose. Shown are the equations required to convert the discrete-time model to its continuous-time equivalent for a single patient, i.e., to match the maximal parasite kill rate (Vmax in equation 3) in the instantaneous model to its equivalent V′max value in the discrete-time model (equation 4), the latter being denoted by the prime symbol. The circumflex or tilde above Vmax indicates whether adjustment has been made for the effects of stage specificity or short half-life, respectively, to compensate for the lack of drug killing in nonsensitive stages and times when the drug is not present during the 48-h (or 96-h) census period.
