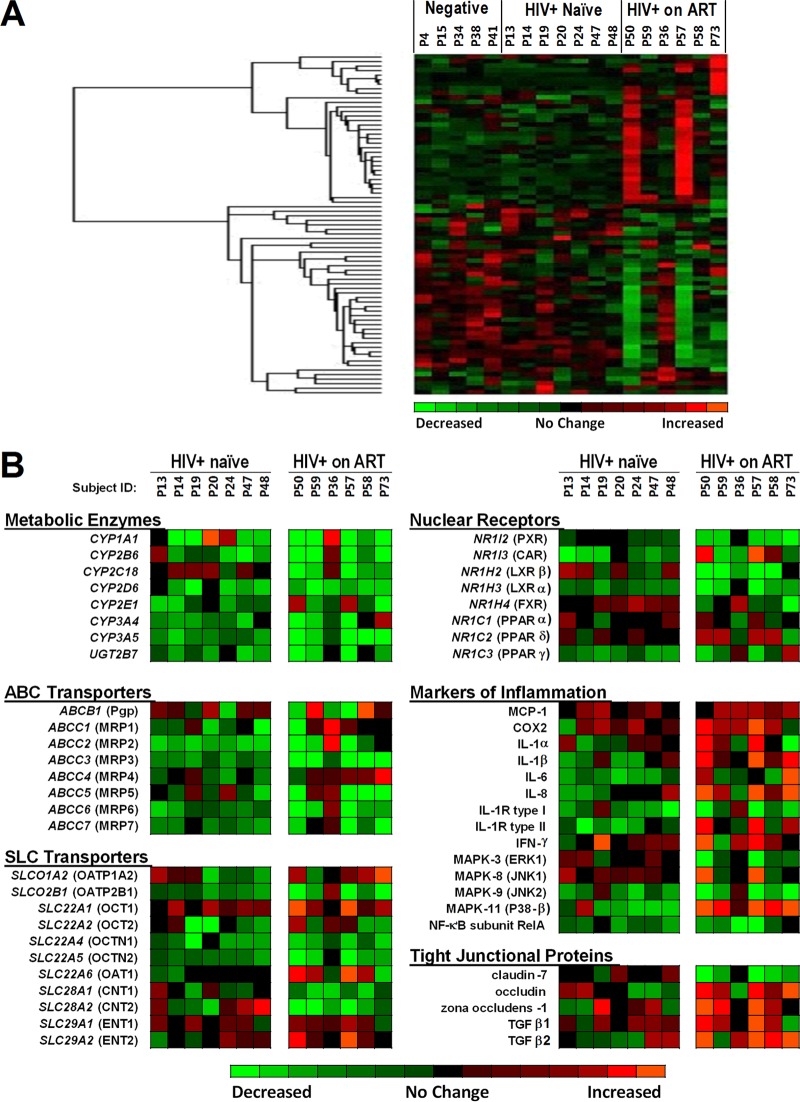
FIG 1.
DNA microarray analysis of mucosal gene expression in treated and untreated HIV-1 infection. (A) Gene expression changes in the gut biopsy specimens of HIV-negative participants (n = 5), HIV+ ART-naive participants (n = 7), and HIV+ participants on ART (n = 6). The patients were arranged based on when the microarray sample was analyzed (see Materials and Methods for details). (B) Differentially expressed genes with important pharmacological and physiological functions, such as genes for metabolic enzymes, drug efflux and uptake transporters (i.e., ABC and SLC drug transporters), nuclear receptors, and tight junctional proteins, and genes associated with the intestinal immune response and inflammation, were identified through hierarchical cluster analysis. For clarity, the sequence of patients was organized by patient identification number matching the arrangement that was used in panel A. Abbreviations: CYP, cytochrome P450 enzyme; UGT, UDP-glucuronyl transferase; Pgp, P-glycoprotein; MRP, multidrug resistance-associated protein; BCRP, breast cancer resistance protein; OATP, organic anion transporting polypeptide; OAT, organic anion transporter; OCT, organic cation transporter; CNT, concentrative nucleoside transporter; ENT, equilibrative nucleoside transporter; PXR, pregnane X receptor; CAR, constitutive androstane receptor; FXR, farnesoid X receptor; LXR, liver X receptor; PPAR, peroxisome proliferator-activated receptor; IL, interleukin; IFN, interferon; MCP, monocyte chemoattractant protein; COX, cyclooxygenase; MAPK, mitogen-activated protein kinase; NF, nuclear factor; TGF, transforming growth factor.