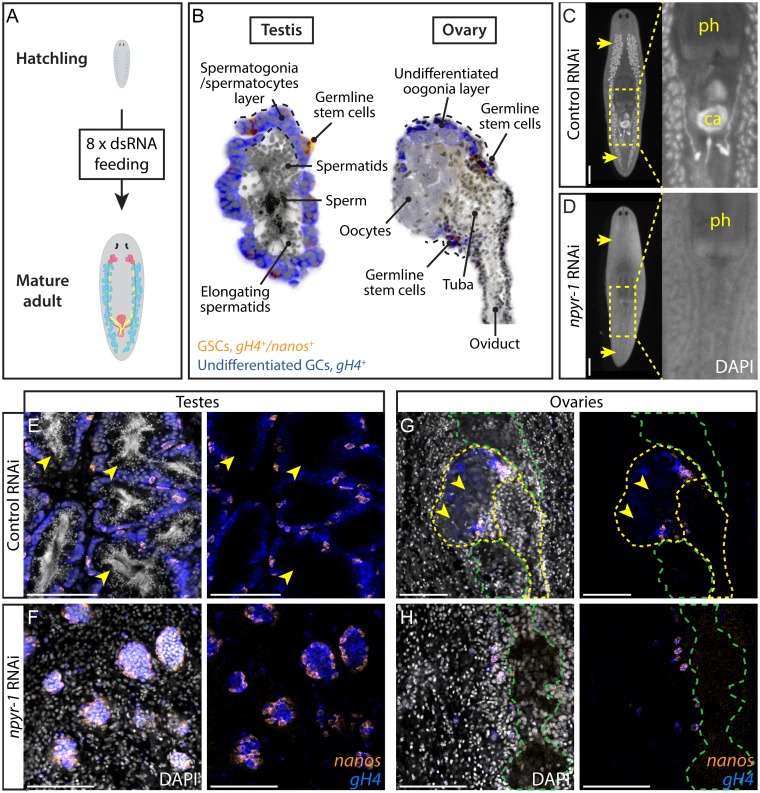
Fig 2. NPY receptor npyr-1 is required for germ cell maturation.
(A) Post-embryonic development RNAi paradigm used to determine the function of genes during normal planarian growth. Hatchlings (≤2 wk old) were fed dsRNA corresponding to each gene eight times to ensure that control worms achieve sexual maturity. (B) Schematic showing planarian testis and ovary structures. In both testes and ovaries, nanos+/gH4+ GSCs (orange) and nanos-/gH4+ spermatogonia/oogonia (blue) are located on the periphery, while more differentiated spermatids, sperm, or oocytes (grey) are in the middle of the gonads. (C, D) DAPI staining showing testes and stored sperm in whole-mount samples. Insets show the copulatory apparatus region. Pharynx and copulatory apparatus are marked by “ph” and “ca”, respectively. RNAi treatment followed the paradigm in A. Control worms (C) develop a complete reproductive system, while npyr-1(RNAi) worms (D) lack developed reproductive tissues and mature gametes. n = 5/5 for each of the three npyr-1 clones and control. RNAi quantification data can be found in S4 Data. (E–H) Double-FISH labeling GSCs (nanos+/gH4+, orange) and spermatogonia (nanos-/gH4+, blue) in whole-mount control and npyr-1(RNAi) samples. GSCs and spermatogonial cells are present in both conditions. Testis germ cells differentiate into sperm and spermatids (arrowheads) in control worms (E), but not in npyr-1(RNAi) worms (F). In ovaries, mature oocytes with large cytoplasm (arrowheads) are surrounded by gH4+ oogonia in control planarians (G), but are absent in npyr-1(RNAi) animals (H). DAPI (grey) labels nuclei. Yellow dashed lines indicate ovaries and oviducts in G and H. Green dashed lines indicate cephalic ganglia near the ovaries. Scale bars are 1 mm in C and D and 100 μm in E–H. See also S3 Fig.