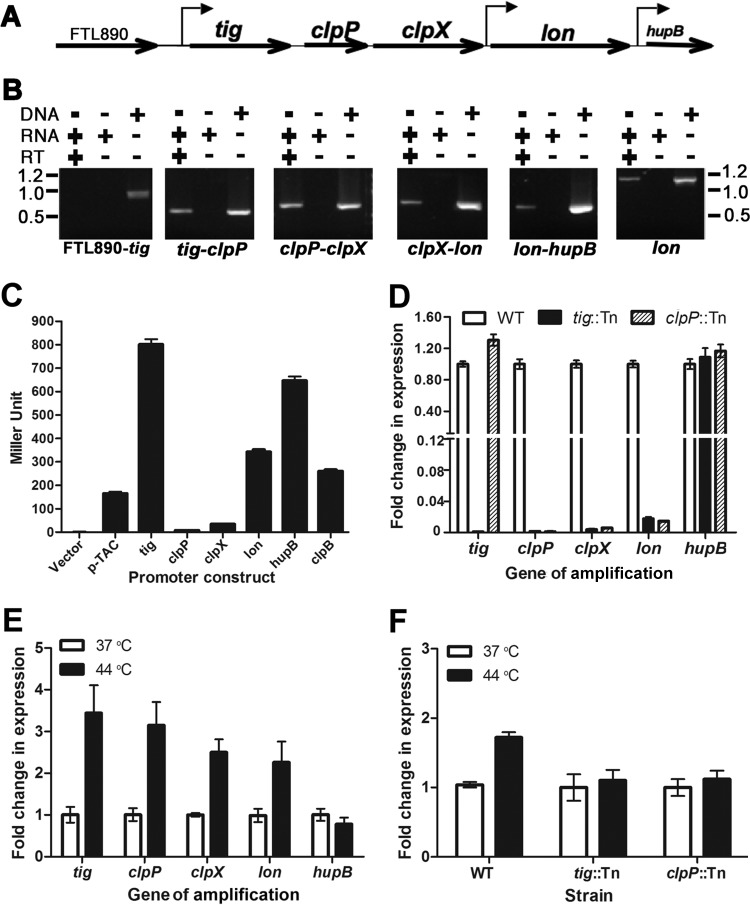
FIG 1.
Transcription of the F. tularensis protease locus. (A) Schematic illustration of the gene arrangement in the protease locus. The gene orientations and promoters identified in panel C are marked by straight and bent arrows, respectively. (B) Cotranscription of the protease locus genes determined by RT-PCR. FTL890-tig, tig-clpP, clpP-clpX, clpX-lon, and lon-hupB junctions were amplified using cDNA, RNA, or DNA as a template. A 1,200-bp internal fragment of lon was amplified as a positive control. The sizes of molecular markers are indicated at the sides of the panel in kilobases. (C) Detection of promoter activity of the 5′ noncoding sequence of each gene in the protease locus with the β-galactosidase reporter. The 5′ noncoding sequence (∼500 bp) immediately upstream of each gene in the protease locus was placed in front of a promoterless β-galactosidase gene in the pMP633 shuttle plasmid. The LVS derivatives containing individual reporter constructs were grown to an OD600 of 0.4 in MHB in the presence of hygromycin (200 μg/ml) at 37°C before being lysed to measure β-galactosidase activity. The constructs for the E. coli pTAC and F. tularensis clpB promoters were used as negative and positive controls, respectively. Bars represent the average β-galactosidase activity ± SEM (n = 3) from one of three independent experiments. (D) Transcription of the protease locus genes in wild-type (WT) LVS and the isogenic tig (tig::Tn) and clpP (clpP::Tn) transposon insertion mutants. The mRNA levels of the protease locus genes, as well as hupB, were quantified by qRT-PCR and normalized on the basis of the DNA helicase gene (FTL1656). The fold changes of the mRNA level in the mutants were converted to percentages of the wild-type value. Bars represent the average ± SEM (n = 3) from one of three independent experiments. (E) Heat-inducible transcription of the protease locus genes in LVS under physiological (37°C) or heat stress (44°C) conditions. The transcripts of the target genes with the total RNA extracts from the LVS culture grown at 37°C or 44°C were quantified and presented as in panel D. (F) Heat-inducible expression transcription of lon from the tig promoter but not from its own promoter. The lon transcripts in LVS (WT) and isogenic tig and clpP transposon insertion mutants were measured with the total RNA extracts from the cultures grown at 37°C or 44°C as for panel D.