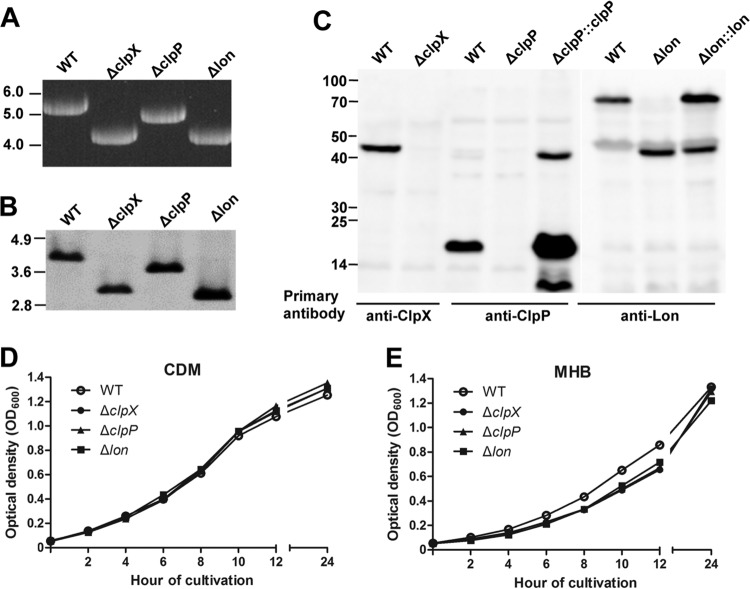
FIG 2.
Verification of unmarked deletions in clpX, clpP, and lon. (A) PCR detection of gene deletions. The protease locus was amplified from genomic DNA of LVS or isogenic clpX, clpP, and lon deletion mutants with the flanking region primers Pr1062/Pr1236. The molecular sizes of DNA markers are marked in kb. (B) Southern blot detection of gene deletions. Genomic DNA preparations of LVS and isogenic ΔclpX, ΔclpP, and Δlon mutants were digested with HindIII and separated by agarose gel electrophoresis. The DNA blot was probed with a PCR fragment (Pr1298/Pr1299) representing a 137-bp coding region of tig. (C) Detection of ClpX, ClpP, and Lon in LVS (WT) or isogenic clpX, clpP, and lon mutants. Cell lysates of each strain (10 μg of total proteins) were separated by SDS-PAGE and tested by immunoblotting with the specific rabbit antiserum for each protein. The sizes of protein standards are indicated at the left in kDa. (D) Growth kinetics of LVS and its derivatives in CDM. The optical densities at 600 nm of LVS (open circles) and the ΔclpX (filled circles), ΔclpP (filled triangles), and Δlon (filled squares) mutants were determined at the indicated time points. The values represent the means ± SEM for three independent cultures from one of the three independent experiments. (E) Growth kinetics of LVS and its derivatives in MHB.